Team:UC-Santa Cruz/Notebook
From 2013.igem.org
| Line 3,317: | Line 3,317: | ||
| - | < | + | <p> |
<strong>Week 2: To create a standard growth curve for CB15</strong> | <strong>Week 2: To create a standard growth curve for CB15</strong> | ||
Revision as of 02:20, 28 September 2013
2013 UCSC IGEM TEAM
Polar Tags Notebook
Kevin Tse and Charles Paines
Groups Goals
We chose to concentrate on four polar tags found in Caulobacter Crescentus. Previous research indicated that DivJ and StpX genes were responsible for polar localization to the stalk side of the cell. Conversely PflI and TipF were chosen as candidates for polar localization to the non-stalk side or flagella side of the cell. These tags were chosen because they appear to be present in the cell during the stalked cell part of the life cycle in Caulobacter Crescentus. The stalked cells with holdfast like stalk structures are ideal for creating biofilms.
Attempts to isolate the polar tag genes from Caulobacter Crescentus involved designing PCR primers which flanked the gene of interest. Restriction sites were also added to the primers to allow for generation of fusion proteins. Generally we used Touchdown PCR cycles to attempt to amplify the polar tags out of the genome. Next the polar tags were ligated into expression vectors which contained either YFP or CFP on the C-terminal side of the multiple cloning sites. These recombinant plasmids were then transformed into competent e.coli cells for plasmid amplification. The amplified plasmids were isolated and sequenced before being transformed back into into Caulobacter.
We also identified a possible minimal (DivJM) polar tag consisting of only 235bp. This minimal DivJ was assembled using overlapping 60 mers synthesized and amplified in a PCR reaction.
General PCR Protocol
For Phusion High-Fidelity PCR Master Mix with GC Buffer:
Initial denaturation:
98°C 30 seconds
25–35 cycles:
98°C 5–10 seconds
45–72°C 10–30 seconds
72°C 15–30 seconds/kb
Final extension:
72°C 5–10 minutes
Hold:
4°C
For OneTaq 2X Master Mix with Standard Buffer:
94°C 30 seconds
30 cycles:
94°C 15–30 seconds
45–68°C 15–60 seconds
68°C 1 minute per kb
Final extension:
68°C 5–10 minutes
Hold:
4-10°C
General Agarose Gel Protocol
Prepare 60 ml of 0.8% agarose solution in TBE buffer by weighing out 0.48 g of SeaKem LE agarose into an Erlenmeyer flask. Add 60 ml of TBE buffer and melt the agarose solution in the microwave. Let the agarose solution cool and pour it into the casting tray and let the agar solidify. Remove comb from the agar. Remove the casting tray from the gel box. Place the gel tray with the gel back into the gel box such that the ends of the gel are pointed into the buffer reservoirs on either end and the end of the gel with wells is pointed towards the negative electrode. Add TBE buffer to the gel box until both reservoirs are full and a 1-2 mm layer of TBE buffer covers the top (flat side facing up) side of the gel. Load samples and DNA ladder on the gel. Slide the lid and connect the electrodes to the power supply. Run the gel for 1 hour at 120 V. After gel run, take a photograph using the gel documentation apparatus.
General Chemicompetent Cell Transformation Protocol
Heat Shock Transformations:
1) Add 100 ul of Top10 Chemicompetent cells in 10 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 200 ul of LB to a microcentrifuge tube.
5) Transfer the Top10 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
LB Media Recipe
Making LB+Kanamycin and LB+Gentamicin Agar Plates:
To make a 500 ml batch:
- 475 ml of MilliQ water
- 5 g of Tryptone
- 5 g of NaCl
- 2.5 g of yeast Extract
- 7.5 g of Agar
We made x2 500 ml batches.
Combine the reagents and shake until the solutes have dissolved. Adjust the ph to 7.0. Sterilize by autoclaving for 20 minutes on liquid cycle.
Take out of the autoclave and let the solution cool. Place in a water bath to prevent solid formation.
Add 1.25 ml of gentamicin antibiotic to one of the 500 ml batches and 500 ul of kanamycin to the other batch.
Pour into petri dishes about half way. Let agar solidify. Place in the refrigerator.
Colony Pick Protocol
Pick a colony using a pipet tip. Place the picked colony in 20 ul of MilliQ water. Pipet up and down to mix the colony thoroughly.
Electroporation Protocol
1) Obtain electrocompetent cells
2) Place sterile electroporator cuvette on ice
3) Turn on the Electroporator and adjust voltage to 1800 V
4) Add 1 ul of the ligation reaction into an aliquot of the electrocompetent cells
5) Electroporate DNA into cells. You want a time constant between 3-5 msec.
6) Pipet 1 ml of LB into cuvette pipeting up and down and transfer back into the 1.5 ml eppendorf microcentrifuge tube.
Reference Sequences
DivJ(full)
1 GAGCTCCTGC GACAGGATTT CCGGAGCAAG CCGATGACGG CGTTCAGAGG GGTTCGCAGT
61 TCGTGGCTGA CCATCCGCAG GAAGGAGCGC TTGCGCTCTT CCAGCGCCAG TTGGCGGCGC
121 GCCTGTGCGG CCGCGCGGCG ATCGGCCGGC GAGGGCGTGA CGGCGCTGTC TTGCGACCGC
181 GTCCTGGATC GGCCGGCTGT GCCATGGGGT CTCCCGGACC TTCTGTCCGT GCTGAATCAG
241 ATGATCAGCA TGGGAGCAGG CGCTTACGAT TCCGTTTCCG TCCACAGGGC TTCCACCACG
301 AAAGGTCGCC CGCAGATTGT GCGGGGTTGG AGGGAAGGCC CGCCCCGCGT TAAGGTTTGA
361 CGACGGTGAT CCTCCCCACC GCGCTAAAAA GTCGACTGGC CTTGGAATTC GAAACGCTTC
421 CAGACCCGTT CAGACGTCCG GCCGCACGCG CCGCCGGGCT CGATCCCGCG CACGCCTGGC
481 GGCTCGGGTG GCTGGCGGCT GTCTGTCTGG CGGCGGCGGC GGCCCTGTTC ACCGCCGACT
541 CCGGCGGTTG GCCTGTGTGG GCGGCGCTCG GCGCCGGCGC CCTGCCCGCC TTGGTCTCGC
601 TGATCTTCAC GCGGGAGGAC GAACGCACCC AGTCCTGGCT TTTGGTGCTG TGGGCCGTGG
661 GCGGCTCGCT GGCGGCGGTG CTGACCGGCG GCGTGGGCGG GGCCATGGCC GCCTGGTGTC
721 TCGCGCCCGT GGCGGCCGCC TCCACTCAGG ATCAGCCCAA GCGTCTGGCT GAGGGCGCGG
781 CCCTGGCCCT GATCGGCGCC TGTGTCGCGG CGCTGACCCA GCTTTCCGGC CTGGCGCCCG
841 CGGCGCCGAC GGGACCGCTG GCCTTCGTTC TGGGCTTTCT GGCGCTGGTG ACGACCGGTC
901 TTGGTCTCGC CGCCGGCCTC CTGATCGGCC GGCGTCGCCA GGGCGCGCGC GACGATCGCT
961 ACGCCAGCGA GATCATCGGT CTGGAGACCT TGCTCGACGG CCTGCCGCAC CTGGCGATCG
1021 CCGTCCGGGG GCAGGGACAG GTGACCGCCG TGCGCGGCGC GGCGCCGCCC GGCGTCACCC
1081 GCGCTGATCT CGTCAATCGC GGTCTGACCG GCGCGGCTGC GCCCGGCGAC CGTCAGCGCC
1141 TGACCGCCGC TATCGCCCAA GCTCATCGTG AGGGCTCGGC CAGCCTGACC TTCAACCCCG
1201 CGCTGGGCGT CGAGCGCGTG GTGGCCCTGG ACATGCACCG CGTTGCGCCG AACCAGCTGG
1261 TCGGCGTGCT GCGCGACATC ACGGTGGAGC GGCATCGCGA GCATGCCCTC GACCAGGCGC
1321 GCATCGACGC CGAGGCCTTG GCCGCCGGCC GCGCGCGGTT CCTAGCCAAT ATGAGCCATG
1381 AGCTTCGCAC GCCCCTGAAC GCCATCATGG GCTTCTCCGA CATCATGCGG GCGCGGATGT
1441 TTGGTCCGCT GAGCGACCGC TACGCCGAGT ACGCTGAGCT GATCCACGAG AGCGGCGGCC
1501 ATCTGCTGGA CCTGATCAAC GACGTGCTGG ACATGTCCAA GATCGAGGCC GAGCGGTTCG
1561 AGCTGCAGCG CGGCGTGTTC GACGCGCGCG AGGCGGTTCA GGCGGCGATG CGGCTGCTGC
1621 GCGTTCAGTC TGACACCGCC GGAGTTCAGC TGCGCGGCGT CCTGCCGCCC GGCGAGCTGG
1681 AGGTCGACGC CGATCGCCGC GCGCTGAAGC AGATCGTCCT GAACCTCGTC TCGAACGCCC
1741 TGAAGTTCAC CCCGCGCGGC GGGCAGGTGA CCGTCACGGC GCACGGCTAT GACGGCGTGC
1801 TCGAGATCGT GGTCGCCGAT ACGGGCGTCG GGATCAGTCC TGAGGACCTG GAACGCCTGG
1861 GGCGTCCCTA CGAGCAGGCC GGCGGGGCCG AACAGCGCGC GCGGGGCACG GGCCTTGGCC
1921 TGTCGCTGGT GCGGGCTTTC GCCCAGCTGC TAGGCGGCGA GATGGTGATC GAGAGCCGCC
1981 TGGGCGCAGG CACGACCGTG TCGGTGCGCC TGCCTGTGCT GCTGGCGCCG ATGGTCGCTG
2041 CCACGCCGAC GCCGCCAGCC GCGCCAGAAG CGCCGTCGGC GCCGGAACCC GCTCCCACGG
2101 TTGAGGAACC GCCGCCGGCC AGCCTGGGCG ACAACGTCAT CGCCTTTGCG CCGCGCTGAG
2161 GCCGCTGGTC TTACCGCAGA ACGCCCGACC GGCGGCGAAG TCTCCGACCT CGATATAGGC
2221 CTTGCGCGTG ACCTCCTCGC CGCGTCCGGC GAAGGCGAAC TCCACCGTGA CCGCTTCACG
2281 AGGGTCCAGT TGCGACATCG CCTCGGCCAC CGCCTCCGGA AACCGGAAGG CCAGGCCCGT
2341 CTTGGCGCCG GTCGGCAGCA GGTGACCGGG GCCTCCTCGC GCCGCTCGGC CG
|
DivJ326
1 GTGATCCTCC CCACCGCGCT AAAAAGTCGA CTGGCCTTGG AATTCGAAAC GCTTCCAGAC
61 CCGTTCAGAC GTCCGGCCGC ACGCGCCGCC GGGCTCGATC CCGCGCACGC CTGGCGGCTC
121 GGGTGGCTGG CGGCTGTCTG TCTGGCGGCG GCGGCGGCCC TGTTCACCGC CGACTCCGGC
181 GGTTGGCCTG TGTGGGCGGC GCTCGGCGCC GGCGCCCTGC CCGCCTTGGT CTCGCTGATC
241 TTCACGCGGG AGGACGAACG CACCCAGTCC TGGCTTTTGG TGCTGTGGGC CGTGGGCGGC
301 TCGCTGGCGG CGGTGCTGAC CGGCGGCGTG GGCGGGGCCA TGGCCGCCTG GTGTCTCGCG
361 CCCGTGGCGG CCGCCTCCAC TCAGGATCAG CCCAAGCGTC TGGCTGAGGG CGCGGCCCTG
421 GCCCTGATCG GCGCCTGTGT CGCGGCGCTG ACCCAGCTTT CCGGCCTGGC GCCCGCGGCG
481 CCGACGGGAC CGCTGGCCTT CGTTCTGGGC TTTCTGGCGC TGGTGACGAC CGGTCTTGGT
541 CTCGCCGCCG GCCTCCTGAT CGGCCGGCGT CGCCAGGGCG CGCGCGACGA TCGCTACGCC
601 AGCGAGATCA TCGGTCTGGA GACCTTGCTC GACGGCCTGC CGCACCTGGC GATCGCCGTC
661 CGGGGGCAGG GACAGGTGAC CGCCGTGCGC GGCGCGGCGC CGCCCGGCGT CACCCGCGCT
721 GATCTCGTCA ATCGCGGTCT GACCGGCGCG GCTGCGCCCG GCGACCGTCA GCGCCTGACC
781 GCCGCTATCG CCCAAGCTCA TCGTGAGGGC TCGGCCAGCC TGACCTTCAA CCCCGCGCTG
841 GGCGTCGAGC GCGTGGTGGC CCTGGACATG CACCGCGTTG CGCCGAACCA GCTGGTCGGC
901 GTGCTGCGCG ACATCACGGT GGAGCGGCAT CGCGAGCATG CGCTCGACCA GGCGCGCATC
961 GACGCCGAGG CCTTGGCC
|
DivJM
1 GCGGCTGCGC CCGGCGACCG TCAGCGCCTG ACCGCCGCTA TCGCCCAAGC TCATCGTGAG
61 GGCTCGGCCA GCCTGACCTT CAACCCCGCG CTGGGCGTCG AGCGCGTGGT GGCCCTGGAC
121 ATGCACCGCG TTGCGCCGAA CCAGCTGGTC GGCGTGCTGC GCGACATCAC GGTGGAGCGG
181 CATCGCGAGC ATGCGCTCGA CCAGGCGCGC ATCGACGCCG AGGCCTTGGC C
|
StpX
1 ATGTTTGGAC GTAATATACG CTTGGCCGCC TTGGCGACCG CCGCGGCAGG CGTTCTTGCG
61 CTGGCGGGCT GTAGTGCGGG CGATCCGGTC GCCTCGGCGC CGCCACCAAG CGATCACCAG
121 AAATCGTCGT CGCCGCCGGA GCTGATGGGC GCGCCGCCGC CCAGCACCGC CCCAGCGCAG
181 GACGGACTTT TGGGCGGGCC GGTCGCTGAC GCCTCGGCGC AGGCCCAGGC TCAGGCGAAA
241 GCCGACGCGG CGCGCCCGCC GCCCCCGTCG CATAACAGCC ACCTGAAGAC CTGGCGCCGT
301 CCGGACGGCA CCCTGGTCAC GGCGATGCGG CCGATCCCCA ATCCCAAGAG CGCGCCGCAC
361 GCCGCCTCGT CGGCGCCCAA GCGCGTCCAC GCCAAGGTCC AGACGCAGGG CTCCTCGGCG
421 CCGGCCAAGC CGGCTGTCGT CGCGGCGGTC AAGCCCGTGA CGCCGAAGAC GGTCGCCCCG
481 GCGCCCGCGC CGGTGAAGGC CGTCCAGCCC GCGCAACCCG CCGCGCCGTT GGCCAAGCCG
541 CCCGTGGTCG CTGCTGCGGC GCCCGCTGCG GCTCCGGCCC CGGCCAAGCC CGCCACCAAG
601 CTTGAACAAC TGCAGGCCGC CGTCGCGCCC GCCGCGACCA ACGGCGCGGT CCTGGCGACC
661 GGCGAAACCC TCGGTAAGGG CCAGCCTGGC CAAGTGACCC TGTCGCTTCC GGCGACCCTG
721 GGCGAGATGA TCCAGAAGGA AGCCGCCAAG CTCGGTTTGG CCAAGGCGGC CAGCAAGACC
781 AGCGCCTATG CCGAACTGCA GGGCGAAGGC TATGAGATCA CCCCGAACGG GCGTCAGACG
841 GCGGTCGTGA AGGCGGGCGA GCCGACCACC TTCGCCTGGC AGGTCAAGCC CGGCGCCGAG
901 GCCAAGGGCG AACTGAAGTC GGAGTTCGGT GTCGAACTGA CCGGCGCGAA GGAACCGCAA
961 GGCTTCTCCC TGGGTGAGAT CAGCAAGCGC GTCGCAGCCC TGCCGCAAGA GGCCAAGAAG
1021 GGCCTCGACA AGCTCGACCT CGGCGCCTTG AACGGCACGA TCTCGCTGCC CGGCCTTGGC
1081 CCGACGCCGA TCAAGACCCT GCTGGGTGCG GCTCTGGTGC TTCTGGCGCT GATCATTCTG
1141 GTGGGCGTGT CGCGCAGCGT CGCCGCTTCG CGCCGCAAGG CCGAAAGCCA GCGCAAGTAC
1201 CGCACGCTGA CCGACTACGG GCGCAACGAG ATGGAGTTCG AGGCGCCTCC GGCCAGCACC
1261 CACGTTTCGC ACGTGAACCC GTTCCTCGCC GCCGCCGGCG GCGCAGCGGC CGGTGCGGTC
1321 GCGGCCTCGG CCGTGGCGCA CCATGACGAT CACCACGACC ACGGGCATGG CCATGATGAT
1381 CACCACGGCC ATGACGACCA CGGCCACGGC CACGATGATC ATGGCCACGA CGCCCACGGT
1441 CATGACGATC ACGGGCACGG GGCGGATCTT CACGCCCACG CCCACCCTGA ACCCCACGCC
1501 CCGGCTGAGC ACGTTTCGTA CGTGAACCCG ATGGTGGCCG CGACCACCGC GAGCCACGCC
1561 GACCACCATG GGCACGATGC TGCTCACGGG CATGATGATC ACGGCCATAG CCACGACGAT
1621 CATGGTCACG GCCACGACGA TCACGGGCAT GGTCACGATG ATCATGCGCA TGGCCACGAC
1681 GATCATGCCC ACGGCCACGA CCATGGCGCC CATCACGCCC ATGCCGAGCA CACCGCCCAT
1741 GTGACCCCGG CCAGCCATGC CGATGATCAC GGCCACGGCC ATGACGACCA TGGACACGGC
1801 CATGACGATC ACGGCCATGA CGCCCATGGT CACGGCGGCC ACGGACATGA TGACCACGGC
1861 CATCACGAAG ACCATGGCCA CGGTCACCAC GACGCGCCCA AAAAGGAACT TGAGCACGCT
1921 CACCAC
|
PflI
1 ATGAGCGTCG TCGCCCTGGC CATGAACGGG TTCCTGGCGG TGCTGCTGAT CGCGGCGCTG
61 ATCTTCGGCT GGCGTCTGGA GCGTCGGCTG AAGGCGCTGC GCGACAGTCA CGAAGGCTTC
121 GCCAAGGCGG TCGCCGATCT CGACCAGGCC GCCGCTCGGG CCGAGCAGGG GCTCGCCGAT
181 CTCCGCGCCG CCACCGACGA GGCCGCTGAA ACCCTGGCCG TCCGGATCGA GCGCGCCCAG
241 GCCCTTGCGG CTCAGCTGGA AGATCGGGTC AACCGGCCTG CGCCGCCCAA GGCTCAGGCA
301 GCGCCCGAAC GCGAGGCTCC GCCACGTCCG TCGCGTGCGA TCGAGGCTCC GCCGGTCCCG
361 CCCCCTGGCG AGCGTCGCCT TTCGGCGGCC GATTTCGAGC GCCTTCTCGA GCGCGAGGAT
421 CGGGTCGAGC GGGCCGCCCG CGGCGAACCG GCGCCGCGAC CCGCGCCACG TCCAAATCTG
481 AGCCAAGAAA CCCCGCGTTC ACGGGCGCGG GTTGATGATG ACCTGTTCGA TGGGCCGGTT
541 GATCCCCCGC GCCCGACCAG CAATCCACGA GTTCCTCGCC GATGAAGAAC ATTC
|
TipF
1 TTGGGACGGA ATGGCGCAGG GACGTTAATC TATTCGACTC TTCAGGCCGG GACCGGTCTG
61 ATTGCGTGTA ACGATTCGTG TGGACGGTTT ATGCGTAGGC TGATGCTGGC GCTCCTGACG
121 GGCGCTTATC TCTGTCTGGC TTTGCTTGTC TCCCTGTTCC TCTTGAGAAC GGGAGCGACT
181 CCGTCCGTGG GCGTCTCGGC GTTCATCGGC ACGTTGGGCC TGTGCTTTGC GTTCCACGGC
241 CTGATCGCCC AAGCGCTGAT GGGCGCGGCC CTGCGCGTCG ACATCGACAC CATCCGCGAA
301 GCCCACGCCA TCCTGCTGGA CCAGATCGAG AAGGTCGACG CCCGCGTCAC CGACCTCGTC
361 GACACCGTCG CCGCCGACGC CCAGCGCCGC TCCGAGGAGC TGTCCAGCGA AGTCCACCAG
421 CTGGAAGACC TGATCCAGCA GATGAACGAC CGCCTGGAGC ACCAGCTTAC CCACCAGGTC
481 GCCGCCGCCT CGGCCCGCAC CGGCGCCCGC GACCGCGCCC CGCAGGCCAG CCACATGCTG
541 CAGGTGGTGC AGGACGCCCT GGCCGAGAAC CGGGTCGACC TCTACCTGCA GCCGATCGTC
601 AGCCTGCCCC AGCGCCGCAC GGTGTTCTAC GAGAGCTTCT CGCGCCTGCG CGATGAGACC
661 GGCCGTGTGA TGATGCCGGC CGAGTATCTG GCGGTGGCCG AGCCCGAGGG CCTGATGACG
721 GCCATCGACA ACCTCTTGCT GTTCCGCTGC GTGCAGATCG TGCGCCGCCT GGCCAAGCAG
781 GACCGCAAGG TCGGGATCTT CTGCAACATC TCGCTGGCCA GCCTGGGCGA CGAGAGCTTC
841 TTCCCGCAGT TCCTGGAGTT CATGCAGGGC AACAAGGACC TGGCCGGGGC GGTGTTCTTC
901 GAGCTGGGCC AGGCGGCGTT CGAGCGTCGC GGTCCGGTCG AGGCCCGCCA CATGGCCCGT
961 CTGGCCAGCC TCGGCTTCAG CTTCAGCCTC GACAAGGTCA ATGACCTGGA CGTCGACTTC
1021 CAGGACCTGG CTCGCGCCGA CGTGAAGTTC CTGAAAGTCG GCGCCCAGAT GATGCTGGAC
1081 CAGCTGGAAG AGCAGGACGG CAAGCTGGTC ATCACCTCGC TGCCCGACCT GAACGCCAGC
1141 GACTTCGCCG CCCTCACCCG CCGCTACGGC ATCGAGGTGA TCGTCGAGAA GGTCGAGGCC
1201 GAGAAGCAGG TCGCCGAGGT GCTGGATCTC GACATCGGCT ATGGCCAGGG CCACCTGTTC
1261 GGCGAGCCGC GCGCCATCCG CGACGCGGTG CTGGCCGAAG CCGATCCGCC CGCCGACTTC
1321 ATCCGCGCCC CGATGCGTCG CCGCGCCGTG GGCTGG
|
References
Lab Notebook 8/6/13
Purpose: To isolate and amplify PflI and Stpx polar markers from Caulobacter Crescentus (CB15N). Expected sizes are 614bp for PflI and 1.9kb for Stpx.
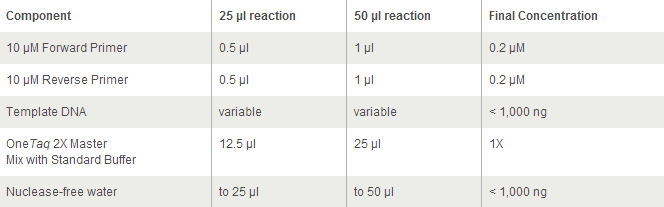
Each PCR sample contains:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15 culture in PYE)
- 0.5 ul of 100 uM forward primer Stpx/PflI
- 0.5 ul of 100 uM reverse primer Stpx/PflI
- 25 ul of OneTaq (2x)
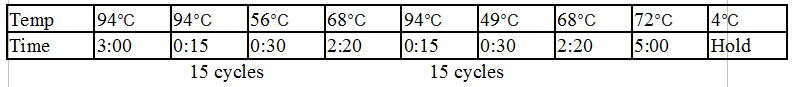
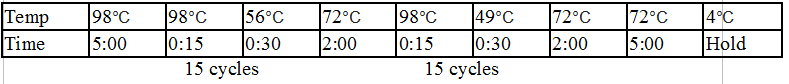
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
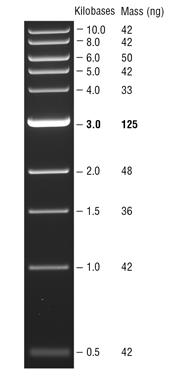
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
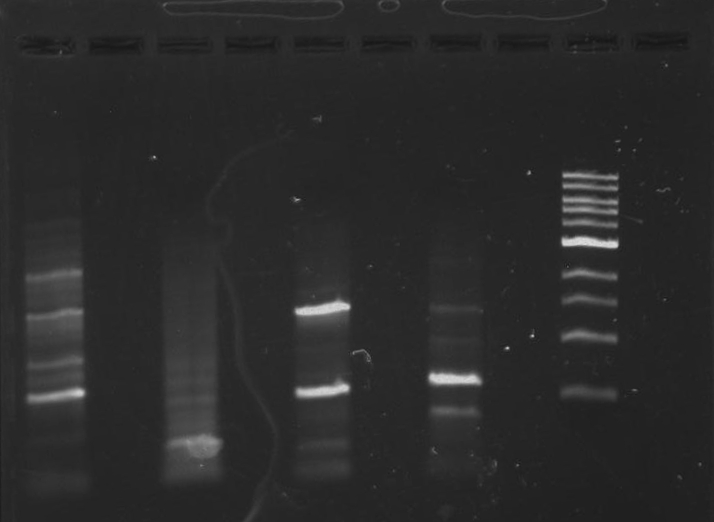
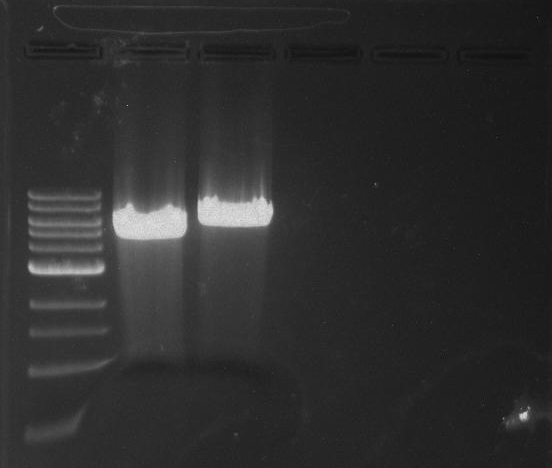
Gel 1: From left to right. PflI, Ladder, Stpx.
Lab Notebook 8/7/13
Each PCR sample consists:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15 culture in PYE)
- 0.5 ul of 100 uM forward primer Stpx/PflI
- 0.5 ul of 100 uM reverse primer Stpx/PflI
- 25 ul of OneTaq (2x)
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 2: From left to right. PflI, Stpx, Ladder.
Lab Notebook 8/8/13
Each PCR sample consists:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15 culture in PYE)
- 0.5 ul of 100 uM forward primer Stpx/PflI
- 0.5 ul of 100 uM reverse primer Stpx/PflI
- 25 ul of OneTaq (2x)
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 3: From left to right. PflI, Stpx, Ladder.
Lab Notebook 8/9/13
Each PCR sample consists:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15 culture in PYE)
- 0.5 ul of 100 uM forward primer Stpx/PflI
- 0.5 ul of 100 uM reverse primer Stpx/PflI
- 25 ul of OneTaq (2x)
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 4: From left to right. Ladder, Stpx, PflI.
Lab Notebook 8/13/13
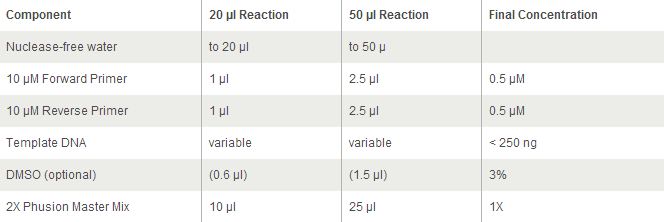
Each PCR sample will contain:
- 9.5 ul MilliQ water
- 1 ul of template (colony pick from CB15N colony pick in PYE)
- 1 ul of forward primer at 100 uM concentration Stpx/PflI
- 1 ul of reverse primer at 100 uM concentration Stpx/PflI
- 12.5 ul of 2x Phusion with GC
PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 5: From left to right. PflI, Stpx, Ladder, PflI, N/A, Stpx, N/A.
Lab Notebook 8/15/13
Two samples of Stpx and PflI were made.
Each PCR sample will contain:
- 23 ul MilliQ water
- 1 ul of template (colony pick from CB15M colony pick in PYE)
- 0.5 ul of PflI/Stpx forward primers at 100 uM concentration
- 0.5 ul of PflI/Stpx reverse primers at 100 uM concentration
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 6: From left to right. Stpx, PflI, Ladder, Stpx, PflI.
Stpx, PflI, TipF, DivJ PCR:
Four samples will contain:
- 2.5 ul forward primer (Stpx, PflI, DivJ, TipF) at 10 uM concentration
- 2.5 ul reverse primer (Stpx, PflI, DivJ, TipF) at 10 uM concentration
- 1 ul template of CB15N made from a colony pick diluted in water
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
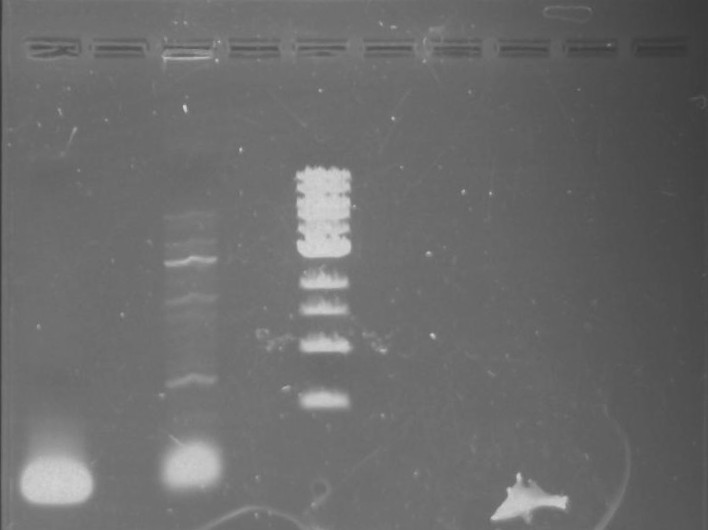
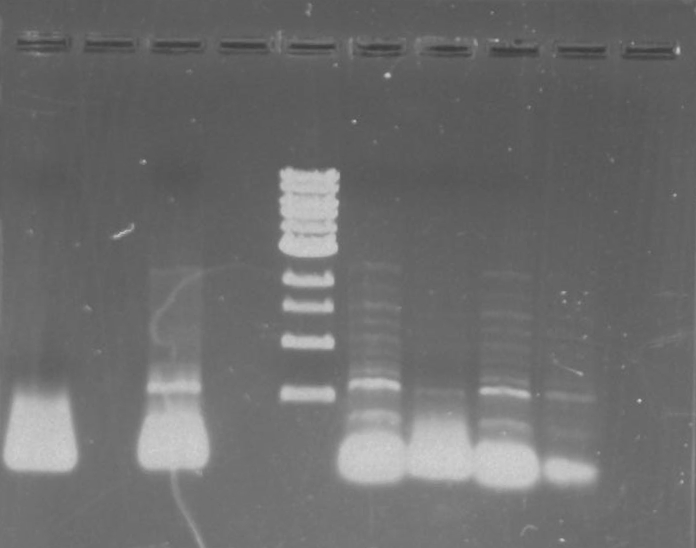
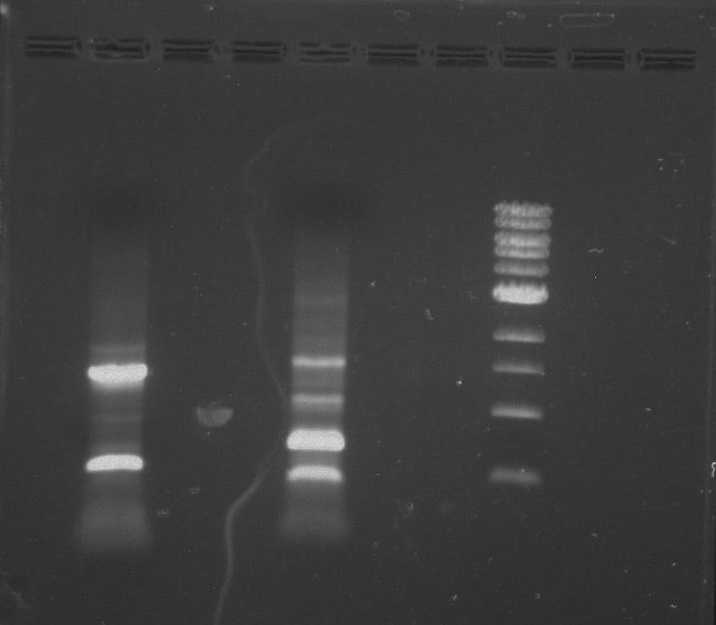
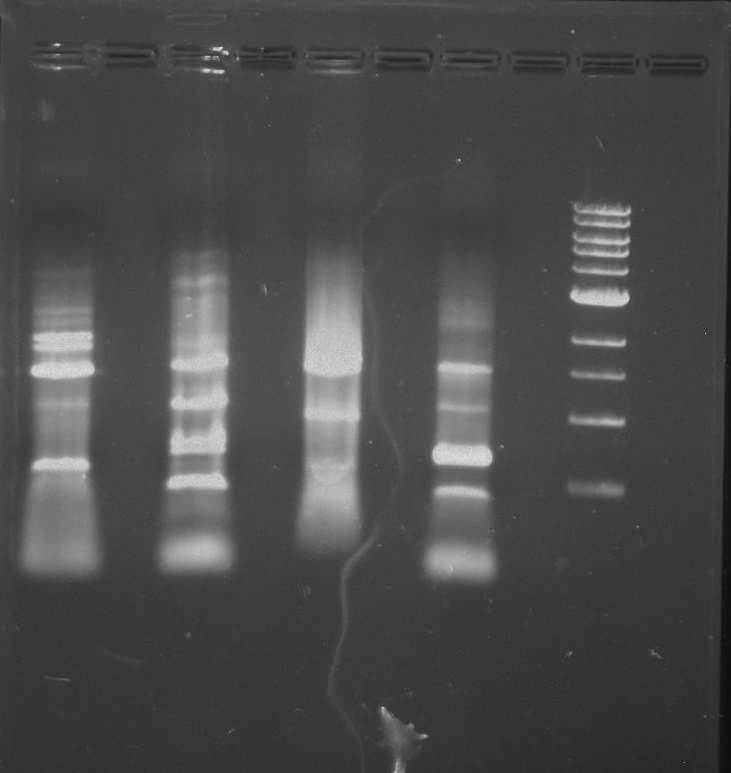
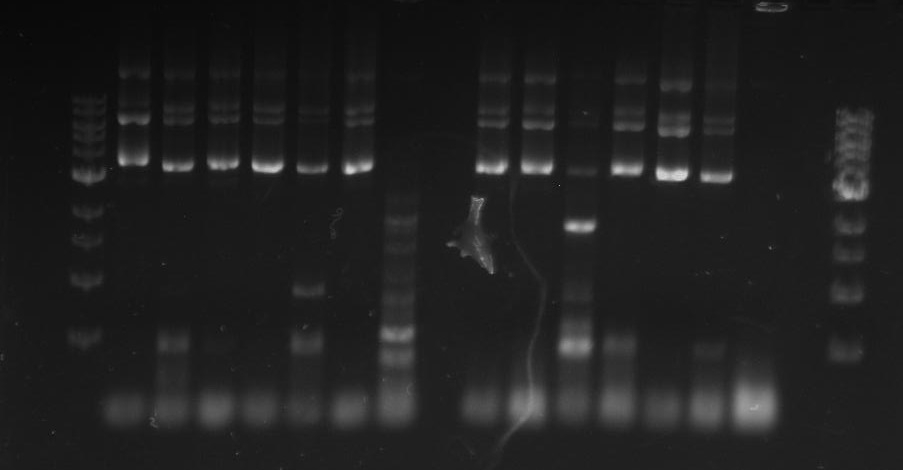
Gel 7: from left to right Stpx, PflI, TipF, DivJ, Ladder.
Lab Notebook 8/16/13
Stpx and TipF PCR:
One Stpx sample and one TipF sample were made.
- 1 ul forward primer (Stpx/TipF) at 10 uM concentration
- 1 ul reverse primer (Stpx/TipF) at 10 uM concentration
- 1 ul template from CB15N glycerol stock
- 9.5 ul MilliQ water
- 12.5 of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 120 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Gel 8: left to right Stpx, ladder, TipF.
Ran the remaining PCR product of TipF and DivJ (45 ul) on a 0.8% agarose gel made from 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 45 ul PCR product + 8 ul 6X gel loading buffer.
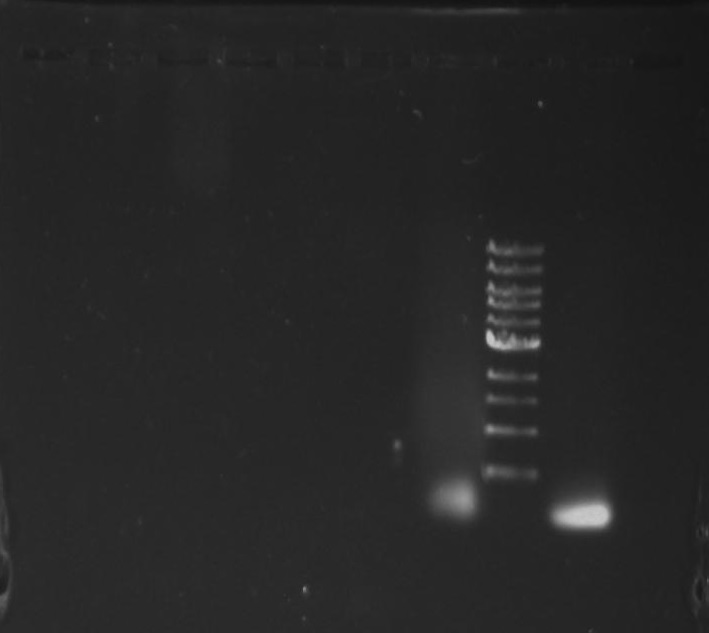
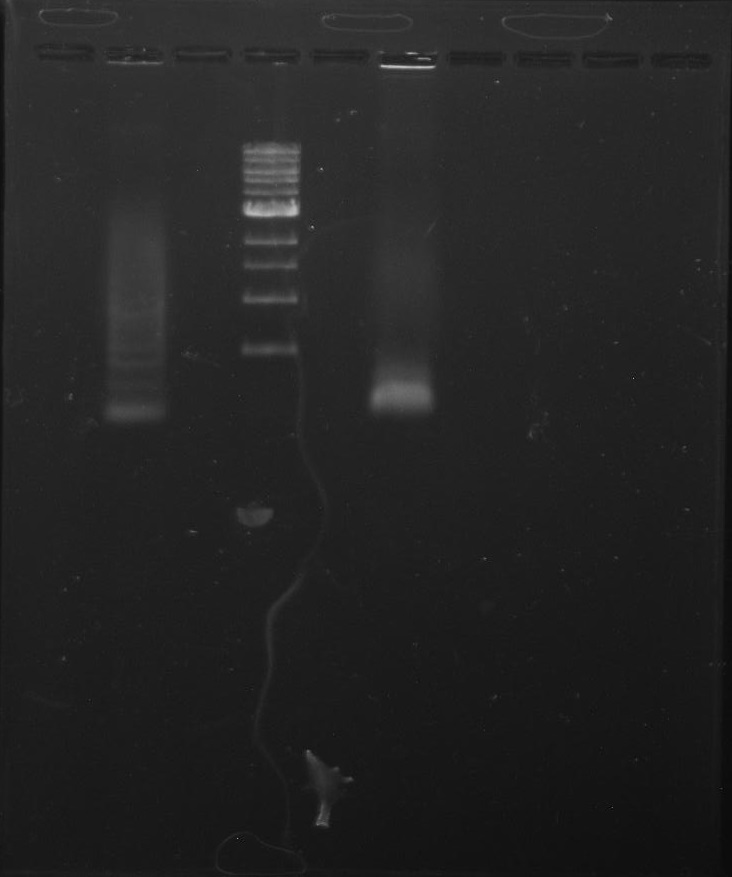
Gel 9: from left to right TipF, DivJ, Ladder.
*gel shows bright bands at expected lengths of ~1.9 kb for TipF and ~600bp for divJ
Purifying DNA from an Agarose Gel:
1) Cut band out in a dark room with a sterile razor and an UV lamp.
2) Place the cut of gel band in a
3) Centrifuge the gel band for 10 minutes at 5,000 xg.
4) Add five volumes of DNA Binding buffer to each volume of DNA sample.
5) Load the mixture into a Zymo-Spin Column in a collection tube.
6) Centrifuge at 10,000 xg for 30 seconds. Discard flow through.
7) Add 200 ul of DNA Wash buffer to the column and centrifuge at 10,000 xg for 30 seconds.
8) Place the Zymo-Spin column into a new 1.5 ml tube.
9) Add 25 ul MilliQ water to column matrix and spin at 10,000 xg for 30 seconds to elute the DNA.
Lab Notebook 8/19/13
PflI and DivJ PCR:
Two samples:
- 1 ul forward primer (PflI/DivJ) at 10 uM concentration
- 1 ul reverse primer (PflI/DivJ) at 10 uM concentration
- 1 ul template of CB15N made from a colony pick diluted in water
- 9.5 ul MilliQ water
- 12.5 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
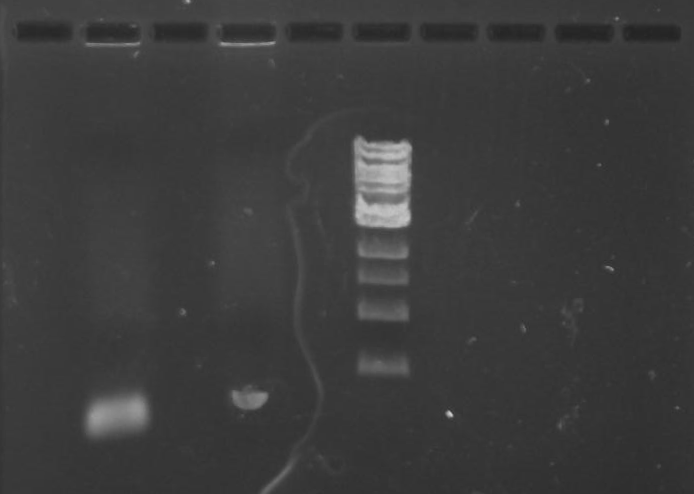
Gel 10: From left to right. PflI, ladder, DivJ.
Redo TipF and DivJ DNA Purification:
Four PCR samples (x2 TipF and x2 DivJ):
- 2.5 ul forward primer (TipF/DivJ) at 10 uM concentration
- 2.5 ul reverse primer (TipF/DivJ) at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
Lab Notebook 8/20/13
Gel of 8/19/13 Redo TipF and DivJ:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
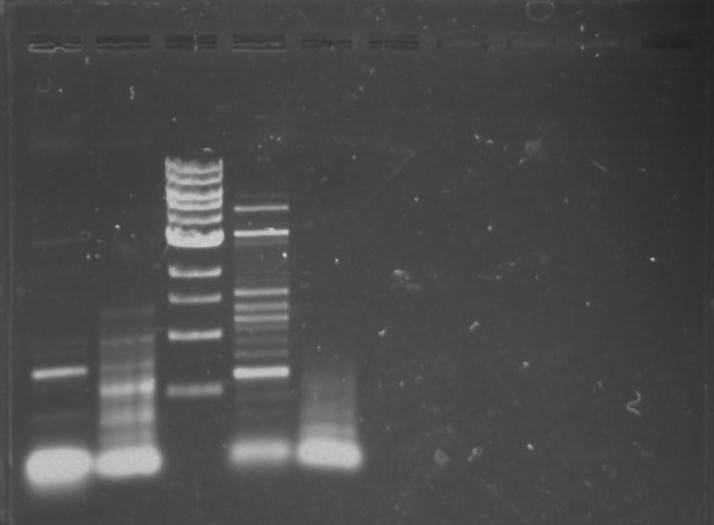
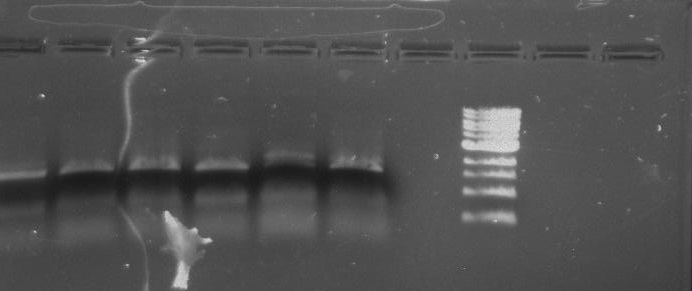
Gel 11: From left to right. TipF, DivJ, TipF, DivJ, Ladder.
Purifying DNA from an Agarose Gel:
1) Cut band out in a dark room with a sterile razor and an UV lamp.
2) Place the cut of gel band in a
3) Centrifuge the gel band for 10 minutes at 5,000 xg.
4) Add five volumes of DNA Binding buffer to each volume of DNA sample.
5) Load the mixture into a Zymo-Spin Column in a collection tube.
6) Centrifuge at 10,000 xg for 30 seconds. Discard flow through.
7) Add 200 ul of DNA Wash buffer to the column and centrifuge at 10,000 xg for 30 seconds.
8) Place the Zymo-Spin column into a new 1.5 ml tube.
9) Add 25 ul MilliQ water to column matrix and spin at 10,000 xg for 30 seconds to elute the DNA.
12 PCR Reactions of DivJ and TipF (6 each):
x6 TipF:
- 2.5 ul forward primer TipF at 10 uM concentration
- 2.5 ul reverse primer TipF at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
x6 DivJ:
- 2.5 ul forward primer DivJ at 10 uM concentration
- 2.5 ul reverse primer DivJ at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
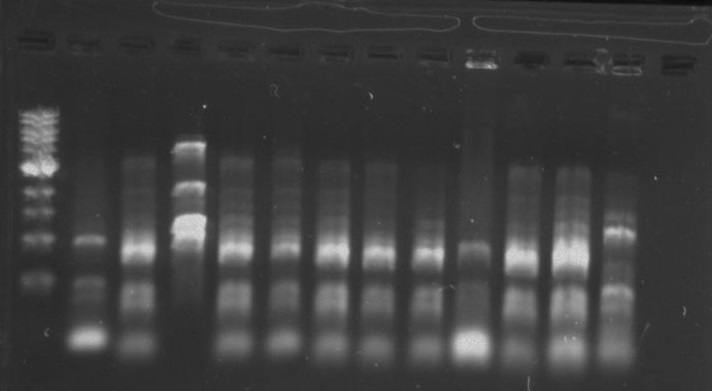
Gel 12: From left to right. TipF, TipF, TipF, TipF, TipF, TipF, Ladder.
Gel 13: From left to right. DivJ, DivJ, DivJ, DivJ, DivJ, DivJ, Ladder.
Lab Notebook 8/21/13
Checking the Purified Samples of TipF and DivJ for One Band:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 5 ul PCR product + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
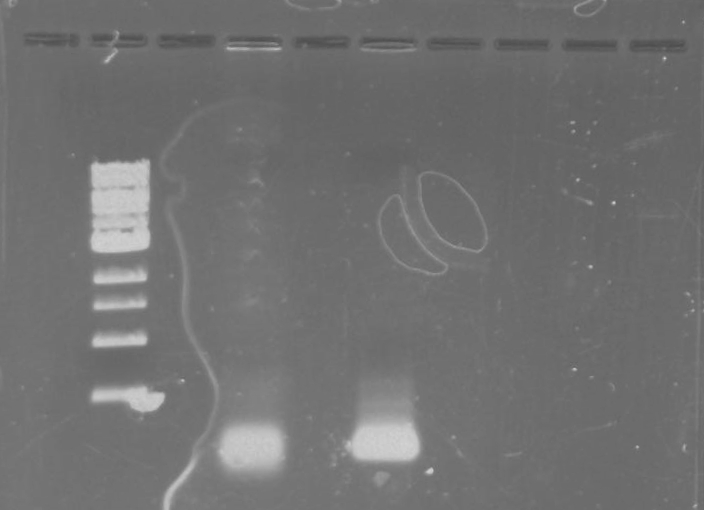
Gel 14: From left to right. TipF, DivJ, Ladder.
x2 TipF:
- 2.5 ul forward primer TipF at 10 uM concentration
- 2.5 ul reverse primer TipF at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
x2 DivJ:
- 2.5 ul forward primer DivJ at 10 uM concentration
- 2.5 ul reverse primer DivJ at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
*Forgot to take a picture of the gel but the gel picture was not good. The TipF samples were not near the expected size. The DivJ samples were smeared.
Lab Notebook 8/22/13
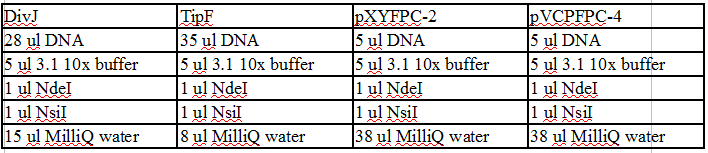
Restriction Digest with NdeI and NsiI
1) Incubate in the PCR thermocycler at 37°C for 30 minutes then at 65°C for 20 minutes to inactivate.
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
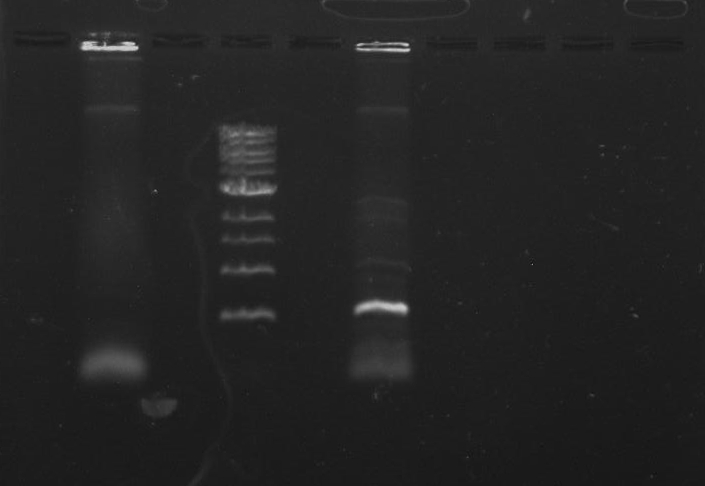
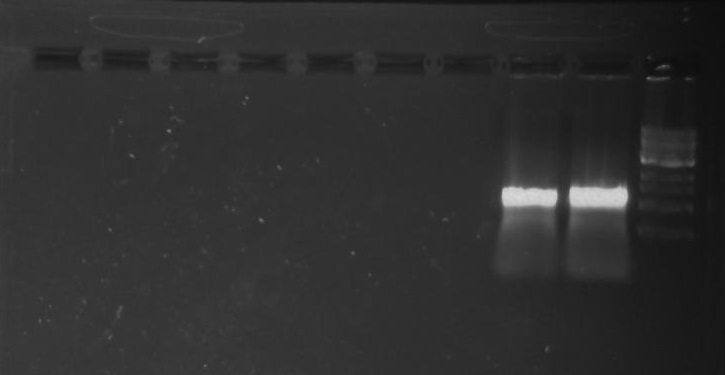
Gel 15: From left to right. Ladder, DivJ with NotI, DivJ without NotI, pXYFPC-2, pVCPFPC-4.
Purifying Vector DNA from an agarose gel
1) Cut band out in a dark room with a sterile razor and an UV lamp.
2) Place the cut of gel band in an agarose spin column.
3) Centrifuge the gel band for 10 minutes at 5,000 xg.
4) Add five volumes of DNA Binding buffer to each volume of DNA sample.
5) Load the mixture into a Zymo-Spin Column in a collection tube.
6) Centrifuge at 10,000 xg for 30 seconds. Discard flow through.
7) Add 200 ul of DNA Wash buffer to the column and centrifuge at 10,000 xg for 30 seconds.
8) Place the Zymo-Spin column into a new 1.5 ml tube.
9) Add 25 ul MilliQ water to column matrix and spin at 10,000 xg for 30 seconds to elute the DNA.
NotI and DivJ Restriction Digest:
- 12.5 ul DivJ DNA
- 5 ul CutSmart 10X buffer
- 1 ul NotI
- 31.5ul MilliQ water
1) Incubate in the PCR thermocycler at 37°C for 30 minutes.
Refer to Gel 15.
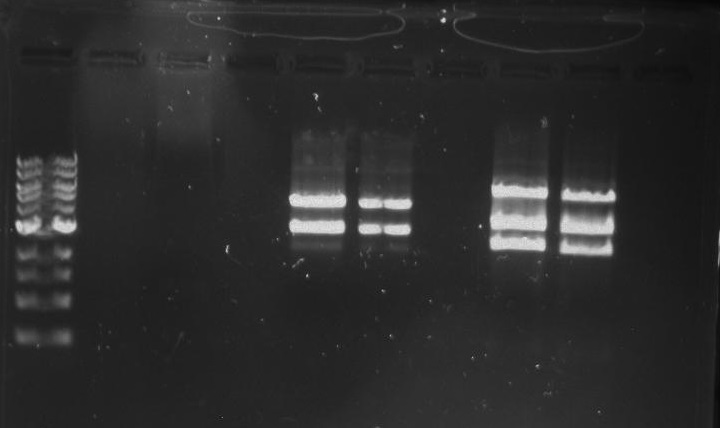
Lab Notebook 8/23/13
DivJ and TipF Ligations
Six total ligation reactions:
Incubate ligation reactions for 30 minutes or greater at room temperature.
Heat Shock Transformations:
1) Add 40 ul of Top10 Chemicompetent cells in 10 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 200 ul of LB to a microcentrifuge tube.
5) Transfer the Top10 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
Plating the Transformations:
1) Obtain three plates of LB and gentamicin and three plates of LB and kanamycin.
2) Warm up the plates in the 37°C incubator prior to plating.
3) Pipet all of the transformation into the plate.
4) Using a bunsen burner, heat up the steel rod.
5) Create streaks with the steel rod.
6) Wait for 5 minutes until all the liquid is soaked up by the plate.
7) Tape up the plates and/or place the plates in a plastic bag.
8) Place plates in an incubator at 37°C.
Lab Notebook 8/25/13
Created an overnight of the DivJ and TipF plates by picking five colonies from each LB and gentamicin plate plus one colony from the negative control. The colony picks were diluted in 20 ul of MilliQ water.
Lab Notebook 8/26/13
Purpose: colony plasmid isolation
Miniprep of TipF and DIvJ:
1) Add 100 ul of 7X Lysis buffer to 600 ul of e. coli culture in a 1.5 ml microcentrifuge tube. Mix by inverting the tube 4-6 times and lyse samples for 1-2 minutes.
2) Add 350 ul of cold Neutralization buffer. Mix thoroughly. Neutralization is complete when sample becomes yellow and precipitate has formed.
3) Centrifuge at 16,000 xg for 2 minutes.
4) Transfer the supernatant into the Zymo-Spin IIN column .
5) Place column into a collection tube and centrifuge at 11,000 xg for 15 seconds. Discard the flow through and place the column back into the same collection tube.
6) Add 200 ul of ENdo Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
7) Add 400 ul of Zyppy Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
8) Transfer the column into a clean 1.5 ml microcentrifuge tube then add 30 ul of MilliQ water. Let stand for one minute at room temperature. Centrifuge at 11,000 xg for 15 seconds to elute the DNA.
Purpose: diagnostic PCR reactions to amplify target sequences
PCR:
x5 TipF
- 1 ul of eluted TipF DNA
- 1 ul of forward primer TipF at 10 uM concentration
- 1 ul of reverse primer TipF at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
x5 DivJ
- 1 ul of eluted DivJ DNA
- 1 ul of forward primer DivJ at 10 uM concentration
- 1 ul of reverse primer DivJ at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
x2 Positive Control
- 1 ul of CB15N glycerol stock
- 1 ul of forward primer DivJ/TipF at 10 uM concentration
- 1 ul of reverse primer DivJ/TipF at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
x2 Negative Control
- 1 ul of colony pick from the negative control plate
- 1 ul of forward primer DivJ/TipF at 10 uM concentration
- 1 ul of reverse primer DivJ/TipF at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
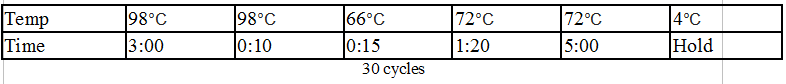
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.46g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
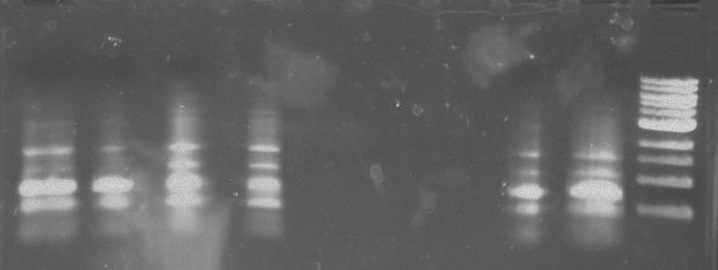
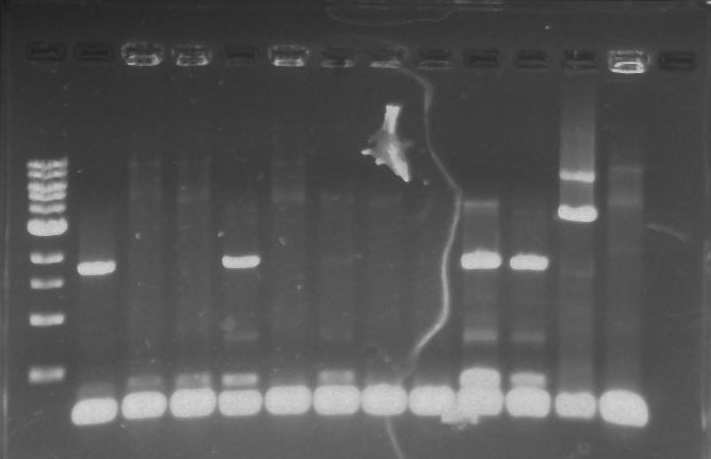
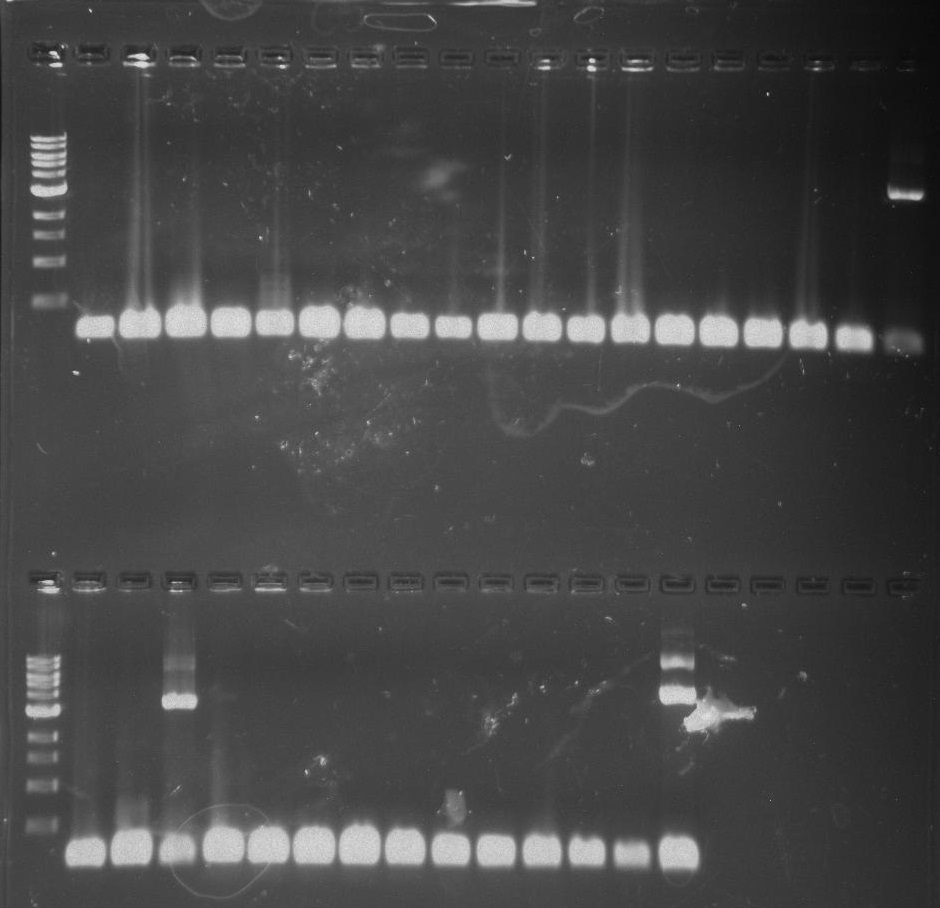
Gel 16: From left to right: Ladder, TipF1, TipF2, TipF3, TipF4, TipF5, Negative Control from TipF plate, Positive Control, DivJ1, DivJ2, DivJ3, DivJ4, DivJ5, Negative Control from DivJ plate, Positive. Control.
Sequencing:
We sent in TipF5 and DivJ3 and DivJ4 in for sequencing.
For TipF5, we used 10 ul of the miniprepped TipF DNA and 3 ul of the forward primer at 1 uM concentration. Also 10 ul of the miniprepped TipF DNA and 3 ul of the reverse primer at 1 uM concentration.
For DivJ3, we used 10 ul of the miniprepped DivJ DNA and 3 ul of the forward primer at 1 uM concentration. Also 10 ul of the miniprepped DivJ DNA and 3 ul of the reverse primer at 1 uM concentration.
For DivJ4, we used 10 ul of the miniprepped DivJ DNA and 3 ul of the forward primer at 1 uM concentration. Also 10 ul of the miniprepped DivJ DNA and 3 ul of the reverse primer at 1 uM concentration.
Lab Notebook 8/27/13
x10 TipF samples:
- 1 ul of eluted miniprepped TipF DNA
- 1 ul of forward primer TipF at 10 uM concentration
- 1 ul of reverse primer TipF at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
x10 DivJ samples:
- 1 ul of eluted miniprepped DivJ DNA
- 1 ul of forward primer DivJ at 10 uM concentration
- 1 ul of reverse primer DivJ at 10 uM concentration
- 2 ul MilliQ water
- 5 ul of 2x phusion mix with GC buffer
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
Gel 17: From left to right. Ladder, TipF1, TipF2, TipF3, TipF4, TipF5,TipF6, TipF7, TipF8, TipF9, TipF10, Negative Control from TipF plate, Positive Control.
*TipF3 shows a band at expected length
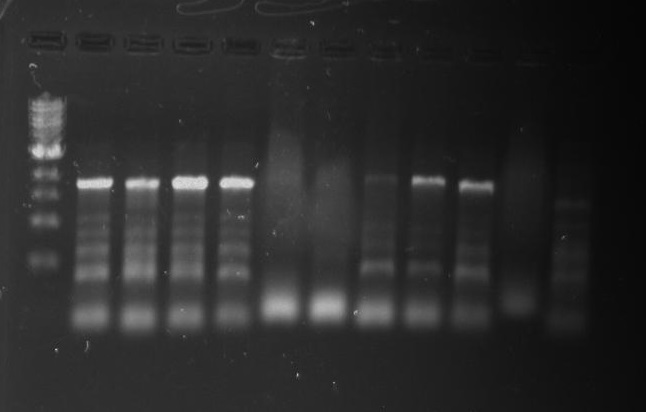
Gel 18: From left to right. Ladder, DivJ1, DivJ2, DivJ3, DivJ4, DivJ5, DivJ6, DivJ7, DivJ8, DivJ9, DivJ10, Negative Control from DivJ plate, Positive. Control.
Overnight culture of TipF3 for Miniprep:
From the colony pick from the LB + gentamicin TipF plate, we diluted it in 20 ul of MilliQ water. That mixture was then transferred into 4 ml of LB. 6 ul of gentamicin antibiotic was added. The overnight culture was stored in the incubator at 28°C at 220 rpm.
Lab Notebook 8/28/13
Miniprep of Overnight TipF3 Culture:
Spin down overnight culture for 10 minutes at 3,000 xg. Discard 3.4 ml of the supernatant. Resuspend the pellet.
1) Add 100 ul of 7X Lysis buffer to 600 ul of e. coli culture in a 1.5 ml microcentrifuge tube. Mix by inverting the tube 4-6 times and lyse samples for 1-2 minutes.
2) Add 350 ul of cold Neutralization buffer. Mix thoroughly. Neutralization is complete when sample becomes yellow and precipitate has formed.
3) Centrifuge at 16,000 xg for 2 minutes.
4) Transfer the supernatant into the Zymo-Spin IIN column .
5) Place column into a collection tube and centrifuge at 11,000 xg for 15 seconds. Discard the flow through and place the column back into the same collection tube.
6) Add 200 ul of ENdo Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
7) Add 400 ul of Zyppy Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
8) Transfer the column into a clean 1.5 ml microcentrifuge tube then add 30 ul of MilliQ water. Let stand for one minute at room temperature. Centrifuge at 11,000 xg for 15 seconds to elute the DNA.
Sequencing of TipF3:
- 10 ul of miniprepped TipF + 3 ul of TipF forward primer at 1 uM concentration
- 10 ul of miniprepped TipF + 3 ul of TipF reverse primer at 1 uM concentration
Lab Notebook 8/29/13
pVan-for and eGYC-1 PCR of TipF Colony Picks:
Mastermix:
- 60 ul of 2x Phusion mix with GC buffer
- 24 ul MilliQ water
- 12 ul pVan-for primer at 10 uM concentration
- 12 ul eGYC-1 primer at 10 uM concentration
Colony Picks:
- Colony pick of TipF in the LB and gentamicin plate into 20 ul of MilliQ water.
Twelve PCR Reactions:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.46g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
Gel 19: From left to right: Ladder, Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer.
Lab Notebook 8/30/13
x2 DivJ samples:
- 2.5 ul forward primer DivJ at 10 uM concentration
- 2.5 ul reverse primer DivJ at 10 uM concentration
- 1 ul template of CB15N glycerol stock
- 19 ul MilliQ water
- 25 ul of 2x Phusion mix with GC buffer
Negative Control:
- 20 ul MilliQ water
- 2.5 ul forward primer DivJ at 10 uM concentration
- 2.5 ul reverse primer DivJ at 10 uM concentration
- 25 ul of 2x Phusion mix with GC buffer
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 20: From left to right. Ladder, DivJ1, Remaining DivJ1, DivJ2.
Lab Notebook 8/31/13
Miniprep of TipF.
Lab Notebook 9/1/13
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
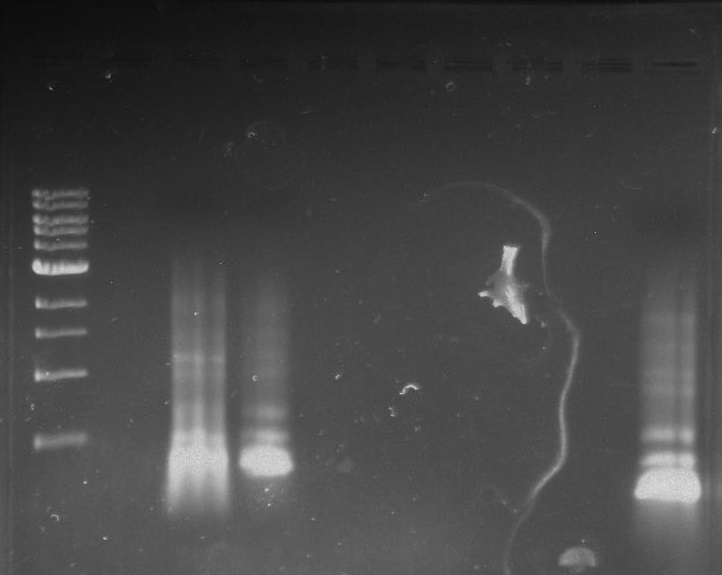
Gel 21: From left to right. TipF, TipF Ladder.
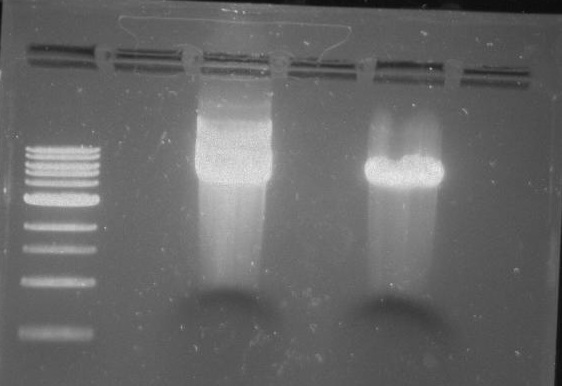
Gel 22: From left to right. Ladder, Vector C, Vector Y.
Cut out the vectors and purify along with DivJ.
Ligation:
Four total ligation reactions:
Heat Shock Transformations:
1) Add 100 ul of BL21 Chemicompetent cells in 10 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 200 ul of LB to a microcentrifuge tube.
5) Transfer the BL21 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
Plating the Transformations:
1) Obtain two plates of LB and gentamicin and three plates of LB and kanamycin.
2) Warm up the plates in the 37°C incubator prior to plating.
3) Pipet all of the transformation into the plate.
4) Add glass beads to the plates and shake.
5) Remove the glass beads and incubate overnight at 37°C.
Lab Notebook 9/2/13
PCR of the 326 DivJ:
Same protocol as 8/30/13
Purified 326 DivJ by cutting it out of the gel and doing a clean and concentrate.
Lab Notebook 9/3/13
Transformation #3
Purpose: To insert DivJ into Vectors and transform ligated fusion vectors/DivJ into competent cells.
Work Flow: Restrict plasmids Y and C. Run restricted plasmids and DivJ(PCR product) on agarose gel. Cut out DNA/ Clean DNA/ Concentrate DNA. Restriction digest of DivJ. Clean and Concentrate DivJ. Ligate sequences. Transform into electrocompetent cells (e.coli top ten).
Restrict plasmids
Vector C
5 uL DNA
5 uL CutSmart (10x) buffer
1 uL NdeI
1 uL AclI
38 uL MilliQ water
Vector Y
5 uL DNA
5 uL CutSmart (10x) buffer
1 uL NdeI
1 uL AclI
38 uL MilliQ water
-incubate at 37° C for 30 min.
Agarose Gel
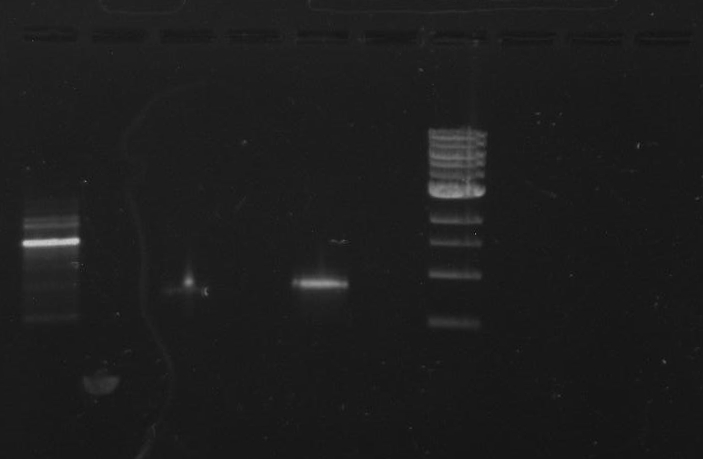
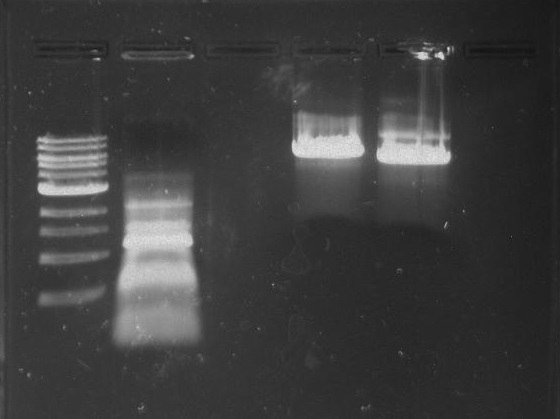
Gel 23: From left to right. 326 DivJ, Vector Y, Vector C.
-cut bands out of gel
-clean/concentrate
-elute vectors to 12 ul
-elute DivJ to 30 ul
Restriction Digest (DivJ)
30 ul DivJ DNA
5 ul CutSmart (10x) buffer
1 ul NdeI
1 ul AclI
13 ul MilliQ water
-incubate at 37° C for 30 min.
-clean/concentrate
-elute to 10 ul
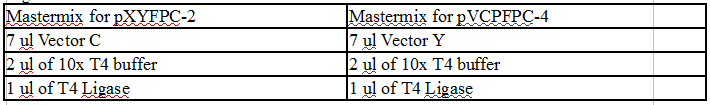
Ligation reactions
Vec C master mix (MMC) Vec Y master mix (MMY)
7 ul vector C 7 ul vector Y
2 ul T4 buffer 2 ul T4 buffer
1 ul T4 ligase 1 ul T4 ligase
-prepare reactions
Neg Control C (NC) Vec C w/ DivJ insert (CD)
5 ul MMC 5 ul MMC
5ul MilliQ H2O 5 ul DivJ
Neg Control Y (NY) Vec Y w/ DivJ insert (NC)
5 ul MMY 5 ul MMY
5 ul MilliQ H2O 5 ul DivJ
- incubate at room temp for 30 min.
Electroporate
Top10 electrocompetent cells
-for each ligation product
250 ul top 10 electrocompetent cells
10 ul ligation reaction
*note too much salt
-recovery in 1 ml LB @ 37° C 1 hour
-plate on LB/kan plates for Y vectors and LB/gen plates for C vectors
Lab Notebook 9/4-5/13
Synthesized the minimal DivJ by:
Creating an oligo mix consisting of 5 ul of each oligo at 100 uM concentration.
Synthesize PCR protocol:
- 1 ul of oligo mix
- 0.5 ul forward primer at 100 uM concentration (NdeI)
- 0.5 ul reverse primer at 100 uM concentration (AciI)
- 23 ul MilliQ water
- 25 ul 2X phusion mix with GC buffer
Amplify PCR Protocol:
- 1 ul of the synthesis PCR product
- 0.5 ul forward primer at 100 uM concentration (NdeI)
- 0.5 ul reverse primer at 100 uM concentration (AciI)
- 23 ul MilliQ water
- 25 ul 2X phusion mix with GC buffer
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
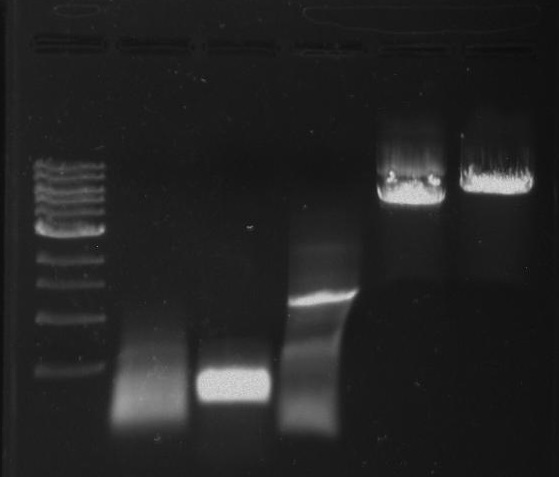
Gel 24: From left to right. Ladder, Synth PCR, Amp PCR, DivJ, Vector C digested, Vector Y digested.
Lab Notebook 9/6/13
Electroporation but this time with BL21 electrocompetent cells.
Plates from this did not look good as well.
* note BL21 was not a good choice for transformations due to the presence of RecA gene.
Lab Notebook 9/7/13
Purpose: prepare top 10 chemi competent cells for use in future transformations.
* note bad results from transformation attempts with top ten electrocompetent cells may have been due to salty ligations and low yields. The chemi competent cells may allow us to use more of our ligation products during transformations.
Making Top10 Chemicompetent Cells:
1) Grow Top10 SOB overnight between the temperatures 20°C and 33°C.
2) Measure the OD and bring the OD back to 0.1.
3) Add 2.5 ml of Top10 SOB to SOB.
4) Incubate at 31°C for one hour and 20 minutes or until the OD is 0.6.
5) Create 5 ml of 1X Washing buffer: 2.5 ml of Dilution buffer and 2.5 ml of Wash buffer (2X)
6) Create 5 ml of 1X Competent buffer: 2.5 ml Dilution buffer and 2.5 ml of Competent buffer (2X)
7) Incubate the culture on ice for ten minutes and then pellet cells at 2,500 xg for ten minutes at 0-4°C.
8) Remove supernatant. Gently resuspend cells in 5 ml of ice cold 1X Wash buffer. Re-pellet as in step 7.
9) Remove supernatant completely. Gently resuspend cells in 5 ml of ice cold 1X Competent buffer.
10) Aliquot 100-200 ul. Flash freeze.
11) Store in -80°C freezer.
Making LB+Kanamycin and LB+Gentamicin Agar Plates:
To make a 500 ml batch:
- 475 ml of MilliQ water
- 5 g of Tryptone
- 5 g of NaCl
- 2.5 g of yeast Extract
- 7.5 g of Agar
We made x2 500 ml batches.
Combine the reagents and shake until the solutes have dissolved. Adjust the ph to 7.0. Sterilize by autoclaving for 20 minutes on liquid cycle.
Take out of the autoclave and let the solution cool. Place in a water bath to prevent solid formation.
Add 1.25 ml of gentamicin antibiotic to one of the 500 ml batches and 500 ul of kanamycin to the other batch.
Pour into petri dishes about half way. Let agar solidify. Place in the refrigerator.
Lab Notebook 9/8/13
Testing the Top10 Chemicompetent Cells:
We tested our Top10 Chemicompetent cells by using different amount of DNA. We used 300 ng of DNA and 30 ng of DNA from our vec C and Vec Y plasmid stocks. The 300 ng of DNA plates had more growth than the 30 ng of DNA plates.
Transformations
K1 G1
1 ul dilute vector Y 1 ul dilute vector C
100 ul CC top 10 cells 100 ul CC top 10 cells
K2 G2
1 ul Vector Y 1 ul Vector C
100 ul CC top 10 cells 100 ul CC top 10 cells
KN GN
1 ul MilliQ H2O 1 ul MilliQ H2O
100 ul CC top 10 cells 100 ul CC top 10 cells
Lab Notebook 9/9/13
Top10 Chemi competent Cell Transformation of DivJ
Heat Shock Transformations:
1) Add 100 ul of Top10 Chemicompetent cells in 10 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 500 ul of SOC to a microcentrifuge tube.
5) Transfer the Top10 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
Colony PCR of TipF
Mastermix:
- 60 ul of 2x OneTaq
- 24 ul MilliQ water
- 12 ul new-pVan-for primer at 10 uM concentration
- 12 ul new-eGYC-1 primer at 10 uM concentration
Colony Picks:
- Colony pick of TipF in the LB and gentamicin plate into 20 ul of MilliQ water.
Twelve PCR Reactions:
Touchdown PCR Protocol:
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
Gel 25: From left to right. Ladder, TipF1, TipF2, TipF3, TipF4, TipF5,TipF6, TipF7, TipF8, TipF9, TipF10, Negative Control from TipF plate, Positive Control.
Lab Notebook 9/10/13
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.8g of agarose and 100 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
Gel 26: From left to right. Top: Ladder, T1, T2, T3, T4, T5, T6, T7, Y8, T9, T10, CD1, CD2, CD3, CD4, CD5, CD6, CD7, NC, PC. Botton: Ladder, CD8, CA1, CA2, CA3, CA4, CA5, CA6, CA7, CA8, CA9, CA10, NC, PC, YD1, YD2, YD3, YD4, NY, PY. T = TipF, CD = 326 DivJ in pVCPFPC-4. CA = Minimal DivJ in pVCPFPC-4. YD = 326 DivJ in pXYFPC-2. NC = Negative control in pVCPFPC-4, PC = Positive control in pVCPFPC-4, NY = negative control in pXYFPC-2, PY = positive control in pXYFPC-2.
Lab Notebook 9/11/13
PCR of 326 DivJ and Minimal DivJ:
326 DivJ:
1 ul template from CB15N glycerol stock
2.5 ul DivJ forward primer at 10 uM concentration (NdeI)
2.5 ul DivJ reverse primer at 10 uM concentration (AciI)
19 ul MilliQ water
25 ul of 2x phusion mix with GC buffer
Minimal DivJ:
1 ul of Synthesis PCR product
0.5 ul DivJ forward primer at 100 uM concentration (248)
0.5 ul DivJ reverse primer at 100 uM concentration (328)
23 ul MilliQ water
25 ul of 2x phusion mix with GC buffer
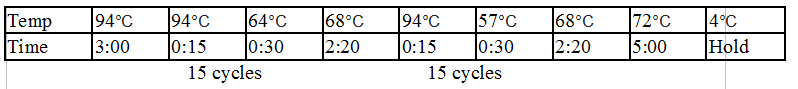
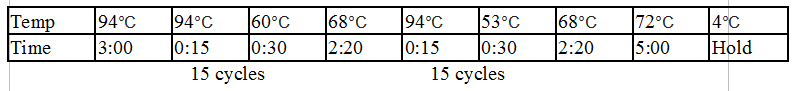
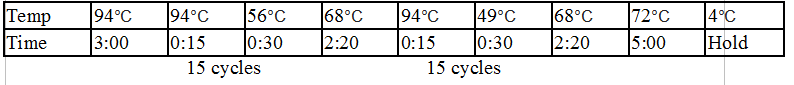
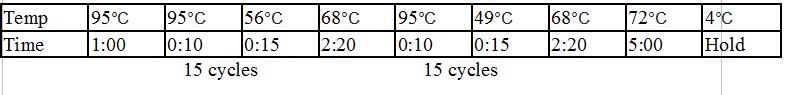
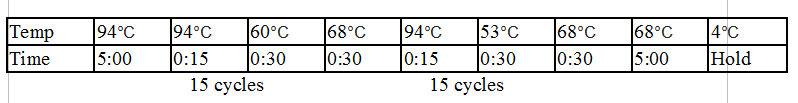
Touchdown PCR Protocol:
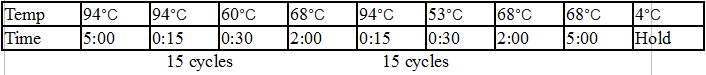
|
Temp |
98°C |
98°C |
64°C |
72°C |
98°C |
57°C |
72°C |
72°C |
4°C |
|
Time |
5:00 |
0:15 |
0:30 |
2:00 |
0:15 |
0:30 |
2:00 |
5:00 |
Hold |
15 cycles 15 cycles
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 27: From left to right. Ladder, 326 DivJ, Minimal DivJ, pXYFPC-2, pYCFPC-4.
-start overnight cultures CA
Lab Notebook 9/12/13
Miniprep of Overnight CA8, CA9, YD1 and YD3 Cultures:
Spin down overnight culture for 10 minutes at 3,000 xg. Discard 3.4 ml of the supernatant. Resuspend the pellet.
1) Add 100 ul of 7X Lysis buffer to 600 ul of e. coli culture in a 1.5 ml microcentrifuge tube. Mix by inverting the tube 4-6 times and lyse samples for 1-2 minutes.
2) Add 350 ul of cold Neutralization buffer. Mix thoroughly. Neutralization is complete when sample becomes yellow and precipitate has formed.
3) Centrifuge at 16,000 xg for 2 minutes.
4) Transfer the supernatant into the Zymo-Spin IIN column .
5) Place column into a collection tube and centrifuge at 11,000 xg for 15 seconds. Discard the flow through and place the column back into the same collection tube.
6) Add 200 ul of Endo Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
7) Add 400 ul of Zyppy Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
8) Transfer the column into a clean 1.5 ml microcentrifuge tube then add 30 ul of MilliQ water. Let stand for one minute at room temperature. Centrifuge at 11,000 xg for 15 seconds to elute the DNA.
Purification of DivJ:
1) Cut band out in a dark room with a sterile razor and an UV lamp.
2) Place the cut of gel band in a
3) Centrifuge the gel band for 10 minutes at 5,000 g.
4) Add five volumes of DNA Binding buffer to each volume of DNA sample.
5) Load the mixture into a Zymo-Spin Column in a collection tube.
6) Centrifuge at 10,000 g for 30 seconds. Discard flow through.
7) Add 200 ul of DNA Wash buffer to the column and centrifuge at 10,000 g for 30 seconds.
8) Place the Zymo-Spin column into a new 1.5 ml tube.
9) Add 12 ul MilliQ water to column matrix and spin at 10,000 g for 30 seconds to elute the DNA.
Restriction Digest of DivJ:
|
Minimal DivJ |
326 DivJ |
pXYFPC-2 |
pYCFPC-4 |
|
28 ul DNA |
28 ul DNA |
3.5 ul DNA |
3.5 ul DNA |
|
5 ul 10X CutSmart buffer |
5 ul 10X CutSmart buffer |
5 ul 10X CutSmart buffer |
5 ul 10X CutSmart buffer |
|
1 ul AciI |
1 ul AciI |
1 ul AciI |
1 ul AciI |
|
1 ul NdeI |
1 ul NdeI |
1 ul NdeI |
1 ul NdeI |
|
15 ul MilliQ water |
15 ul MilliQ water |
39.5 ul MilliQ water |
39.5 ul MilliQ water |
Incubate at 37°C for 30 minutes.
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.4g of agarose and 50 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 50 ul PCR product + 10 ul 6X gel loading buffer.
Gel 28: From left ro right. Ladder, pXYFPC-2, pYCFPC-4.
Lab Notebook 9/13/13
Clean and concentrated DivJ and eluted it to 12 ul. Cut out the vectors. Clean and concentrated and eluted it to 15 ul each.
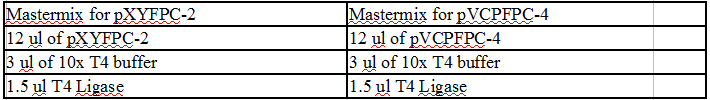
Ligation of DivJ:
|
Mastermix of pXYFPC-2 |
Mastermix of pYCFPC-4 |
|
12 ul pXYFPC-2 |
12 ul pYCFPC-4 |
|
3 ul 10X T4 buffer |
3 ul 10X T4 buffer |
|
1.5 ul T4 Ligase |
1.5 ul T4 Ligase |
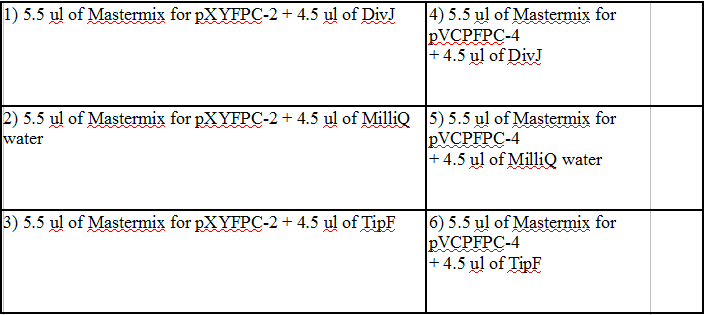
Six total ligation reactions:
|
1) 5.5 ul of Mastermix for pXYFPC-2 + 4.5 ul of 326 DivJ |
4) 5.5 ul of Mastermix for pVCPFPC-4 + 4.5 ul of 326 DivJ |
|
2) 5.5 ul of Mastermix for pXYFPC-2 + 4.5 ul of MilliQ water |
5) 5.5 ul of Mastermix for pVCPFPC-4 + 4.5 ul of MilliQ water |
|
3) 5.5 ul of Mastermix for pXYFPC-2 + 4.5 ul of Minimal DivJ |
6) 5.5 ul of Mastermix for pVCPFPC-4 + 4.5 ul of Minimal DivJ |
Incubate ligation reactions for 30 minutes or greater at room temperature.
Heat Shock Transformations:
1) Add 100 ul of Top10 Chemicompetent cells in 1-2 ul of each ligation reaction.
2) Place in a 42°C water bath for 30 seconds.
3) Place back on ice for 5 minutes.
4) Add 500 ul of SOC to a microcentrifuge tube.
5) Transfer the Top10 Chemicompetent cells and ligation reaction to the microcentrifuge tube.
6) Incubate for one hour at 37°C at 200 rpm.
Plating the Transformations:
1) Obtain three plates of LB and gentamicin and three plates of LB and kanamycin.
2) Warm up the plates in the 37°C prior to plating.
3) Pipet all of the transformation into the plate.
4) Using a bunsen burner, heat up the steel rod.
5) Create streaks with the steel rod.
6) Wait for 5 minutes until all the liquid is soaked up by the plate.
7) Tape up the plates and/or place the plates in a plastic bag.
8) Place plates in an incubator at 37°C.
Lab Notebook 9/14/13
Colony PCR of TipF and DivJ Plates:
Mastermix for pVCPFPC-4 plasmids:
- 110 ul 2x OneTaq
- 44 ul MilliQ water
- 22 ul Pvan-for primer at 10 uM concentration
- 22 ul eGYC-1 primer at 10 uM concentration
Mastermix for pXYFPC-2 plasmids:
- 55 ul 2x OneTaq
- 22 ul MilliQ water
- 11 ul Pxyl-for primer at 10 uM concentration
- 11 ul eGYC-1 primer at 10 uM concentration
Gel Protocol: We created an 0.8% agarose gel by obtaining 0.48g of agarose and 60 ml of 1X TBE buffer. Run at 100 volts for 60 minutes.
Ladder consists of 5 ul DNA Standard + 2 ul 6X gel loading buffer + 5 ul MilliQ water.
Samples consist of 10 ul PCR product + 2 ul 6X gel loading buffer.
Gel 29: From left to right. Top: CD1, CD2, CD3, CD4,CD5, CD6, CD7, CD8, CD9, CD10 CA1, CA2, CA3, CA4, CA5, CA6, CA7, CA8, Positive Control for pVCPFPC-4. Bottom: CA10, Positive Control for pVCPFPC-4, YA1, YA2, YA3, YA4, YA5, YA6, YA7, YA8. YA9, YA10, Positive Control for pXYFPC-2. CD = 326 DivJ in pVCPFPC-4. CA = Minimal DivJ in pVCPFPC-4. YA = Minimal DivJ in pXYFPC-2.
Lab Notebook 9/15/13
Overnight Cultures of YA1, YA2, YA3, YA4, YA5, CA5, CA10, CD2, CD3:
For each overnight:
3 ml of SOC. For pVCPFPC-4, add in 4.5 ul of gentamicin. For pXYFPC-2, add in 1.8 ul of kanamycin.
Lab Notebook 9/16/13
Miniprep of Overnight Cultures:
1) Add 100 ul of 7X Lysis buffer to 600 ul of e. coli culture in a 1.5 ml microcentrifuge tube. Mix by inverting the tube 4-6 times and lyse samples for 1-2 minutes.
2) Add 350 ul of cold Neutralization buffer. Mix thoroughly. Neutralization is complete when sample becomes yellow and precipitate has formed.
3) Centrifuge at 16,000 xg for 2 minutes.
4) Transfer the supernatant into the Zymo-Spin IIN column .
5) Place column into a collection tube and centrifuge at 11,000 xg for 15 seconds. Discard the flow through and place the column back into the same collection tube.
6) Add 200 ul of ENdo Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
7) Add 400 ul of Zyppy Wash buffer to the column. Centrifuge at 11,000 xg for 30 seconds.
8) Transfer the column into a clean 1.5 ml microcentrifuge tube then add 30 ul of MilliQ water. Let stand for one minute at room temperature. Centrifuge at 11,000 xg for 15 seconds to elute the DNA.
Nine Sequencing Samples:
YA1, YA2, YA3, YA4, YA5, CA5, CA10, CD2, CD3
10 ul of miniprepped DNA plasmid with 3 ul of the resepective forward/reverse primer at 1 uM concentration.
Lab Notebook 9/17/13
Made more gentamicin and kanamycin plates.
Re-did the synthesis of the minimal DivJ 60 mers (sequencing results suggest the first DivJM assembly resulted in a single base deletion.
Transformation of the CA8 and CA9 miniprep plasmid that we sent in for sequencing. Changed the amount of SOC added. This time we used 250 ul.
Lab Notebook 9/18/13
Re-did the restriction digest of the vectors and the minimal DivJ with the restriction enzymes NdeI and AciI.
Lab Notebook 9/19/13
Re-did the colony PCR of the TipF plates one last time. Same settings as before.
Lab Notebook 9/20/13
Colony PCR of the Minimal DivJ Plate:
Mastermix for pVCPFPC-4 plasmids:
- 60 ul 2x OneTaq
- 24 ul MilliQ water
- 12 ul Pvan-for primer at 10 uM concentration
- 12 ul eGYC-1 primer at 10 uM concentration
Mastermix for pXYFPC-2 plasmids:
- 60 ul 2x OneTaq
- 24 ul MilliQ water
- 12 ul Pxyl-for primer at 10 uM concentration
- 12 ul eGYC-1 primer at 10 uM concentration
Touchdown PCR Protocol:
2013 UCSC IGEM TEAM Biofilm Membrane Notebook.
The primary goal of the Biofilm Membrane team was to produce a high-density monolayer biofilm. A monolayer biofilm structure is critical to the function of the biomachine as the overall goal of the project is to move salt across a membrane, producing potable water. Caulobacter Crescentus (C. crescentus), is a polar body bacteria that forms high density monolayer biofilms and has been well characterized to support its use in biotechnological applications. The biofilm forming capabilities of a strain of C. crescentus (CB15) have been well characerized by a group at Stanford University whose work we have cited. CB15 was obtained from the Standford lab for the purpose of this project.
Week 1: To prepare materials that will be used for future experiment.
8/15
Peptone Yeast Extract (PYE) Protocol
(for preparation of one Liter)
Aim : To make 1 Liter of PYE growth medium for Caulobacter
Materials :
Bacto Peptone 0.2% approx. 2g lot# 1194403
Yeast Extract 0.1% approx. 1g lot# 1307388 vat# 288620
MgSO4 1mM 0.247g EMD#MX0070-1 lot# uk09FZEMS
CaCl2 o.5mM 0.074g lot# A0141702020
1 Liter flask
milliHQ water
autoclave
Procedure :
1) Combine 2g of bacto peptone, 1g of yeast extract, 0.24624g of MgSO4, and 0.07350g of CaCl2 in a sterile 1 Liter flask.
2) Fill flask with millliHQ water to 1 Liter mark on flask.
3) Homogenize mixture by shaking/stiring.
4) Sterilize mixture using the autoclave on fluid cycle for 40 minutes.
5) Let cool, cover, and store in refrigerator.
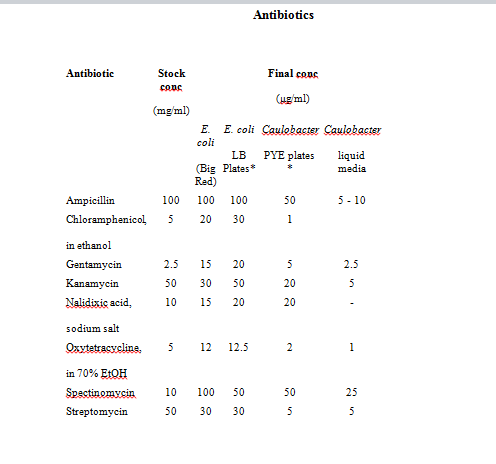
Refer to <a href="http://labs.mcdb.lsa.umich.edu/labs/maddock/protocols/media/antibiotics.html"> </a> for Antibiotics Concentations.
4/9/93
Notes:
Storage: All antibiotics should be stored at 4° C, Tet should be stored at -20° C.
Tet is also light sensitive. Store stock solutions & plates in the dark.
Prep: All antibiotics should be dissolved in sterile distilled water unless otherwise indicated.
Use oxytetracycline for Caulobacter strains.
8/15
Innoculating Ecoli PxYFPC-2
Aim: Picking Ecoli from a plate to get PxYPFC-2 plasmids
Materials:
-Ecoli PxYFPC-2 plate from Stanford
-LB growth medium (made by Aaron)
-Kanamycin ( stock conc. 50mg/ml)
-Inoculating Loop
-Flame
Procedure:
1. Set up a flame
2. Sterilize the loop by heating it over the flame
3. Using the inoculating loop, pick a colony
4. Dip loop into sterile LB broth (100ml)
5. Store in shaking incubator at -37oC for 1 night
8/19 Agar Plates
Aim: To make PYE + antibiotic agar plates
Materials:
-Petri dishes
-PYE (500ml- this is enough to make 15-18 plates)
-Kanamycin/Gentamycin
-Agar (13-15 g per liter) [Lot # 2107450]
-1 Liter Erlenmeyer Flask
Procedure:
1. Add 7.5g of agar and 500mL of PYE to a 1L flask.
2. Homogenize mixture using a stirring bar.
3. Autoclave for 40 mins on fluid cycle.
4. Cool mixture to 50°C
5. add antibiotic to mixture:
6. For antibiotic concentrations refer to:
“A comprehensive set of plasmids for vanillate and xylose-inducible gene expression in Caulobacter Cresentus”
for kanamycin plates: add 250uL kanamycin
for gentamycin plates: add 250uL gentamycin
7. Pour agar into sterilized petri dishes.
8. Let stand for 15 mins to allow agar to solidify.
9. Cover with parafilm and store in refrigerator.
Week 2: To create a standard growth curve for CB15
8/26
Ampicillin dilution
Aim: to make stock ampicillin 50mg/ml
Materials:
-Ampicillin [Lot # 29371]
-BD syringe 0.2micron filtuer
-50ml centrifuge tube
-eppendorf tubes
Procedure:
1. Weight out 0.5 g of Ampicillin
2. Place in 50ml centrifuge tube along w/ 10ml of dH2O.
3. Vortex the solution
4. Using BD syringe filter out 10ml of diluted antibiotic into eppendorf tubes.
5. Fill 20 eppendorf tubes w/ aliquot of 500ul.
8/28
Growth Curve
Aim : to create a standard caulobacter growth curve
Materials:
-PYE+AMP
-50ml centrifuge tubes x6
-CB15
Procedure:
For 33 degrees C
1. Label 1 centrifuge tube “negative” and the other two c tubes. “33 ” and “33 b”
2. Make 80ml of pye + amp.
Final conc of amp = 10ug/ml
Add 80ml of pye into a flask w/ 16ul of ampicillin( stock conc. 50mg/ml)
3. Fill each tube w/ 4ml PYE
4. For negative control tube dip a sterile pipette tip in the tube. Discard tip.
5. For the other 2 tubes pick a colony from CB15 plate w/ a pipette tip. Pick two colonies one for each.
6. Store in incubator at 33C at 300rpm
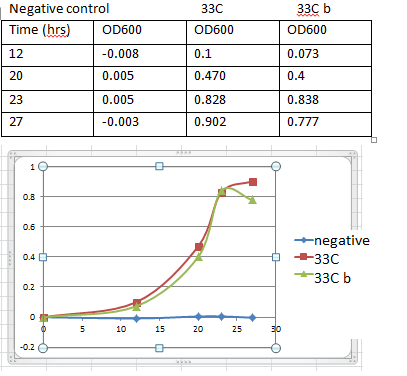
7. Make a table and later transfer in unto a plot of O.D600 versus time graph.
8. Record an OD reading every couple of hours.
Results
8/26
Inoculation of CB15
Aim: making liquid culture of CB15
Materials:
-CB15(from Stanford)
-Antibiotic
-PYE
Procedure:
1. Add 50ul of Ampicillin (50mg/ml) into 250ml of PYE
Refer to for final concentration of Ampicillin(10ug/ml):
“Dynamics and Control of Biofilms of the Oligotrophic Bacterium Caulobacter Crecentus.”
2. Pipet 20ml of pye + amp into centrifuge tube. Do this twice. Label one “Negative” and the other “CB15”
3. Pick a colony from CB15 plate and eject tip in tube
4. For negative control, drop a blank pipet tip
5. Incubate at 28°C and 180 rpm
Results:
After 1 night PYE medium for negative control turned cloudy. Experiment has been contaminated.
8/29
Making Glycerol Stocks
Aim: for long term storage of CB15
Materials:
-glycerol 60% (filtered)
-cell culture cb15
-eppendorf tubes.
Procedure:
1. Prepare a preculuture: Pick a CB15 colony and inoculate it in 5ml of PYE. When culture reaches stationary phase the O.D600 should be around 1.3-1.6
2. Caulobacters are store in 10% glycerol
Refer to: “Caulobacter S-Layer Secretion Kit Manual”
3. Calculate 10% glycerol.
(x amount of 10% glycerol )(60)= (900ul of cells + x)(10)
This comes out to be 180ul of 60% glycerol per 900ul of cb15
4. In an eppendorf tube combine 180ul of 60% glycerol and 900ul of cb15 culture.
5. Shake tube.
6. Label and store tubes at -80C
8/29-8/30
Preparing Electrocompetent CB15
Aim : making electrocompetent CB15
Materials:
-PYE
-eppendorf tubes
-centrifuge tubes(50ml)
-1L falsk
-ice-cold, sterile, distilled water
-ice-cold, sterile 10% glycerol in distilled water.
Procedure : will yield enough electro competent cells for 15 transformation.
1. Diluting 60% stock conc. Glycerol into 10%
Add 80ml of distilled water and 16ml of 60% filtered glycerol from fridge into a flask
2. Autoclave 1000ml of distilled water, 250ml PYE flask (x2), 96ml of 10% glycerol - fluid cycle 40mins
3. Inoculate a CB15 colony into 5ml PYE medium. Grow at 30oC w/ shaking until the O. D is 0.5-1.0 ( should take about 1 night)
4. Record the O.D of the culture above. 0.902
5. Dilution calculation: calculate the amount of culture you would add to 250ml of fresh PYE to achieve an O.D of 0.4-0.6 in 18hrs.
-doubling time of Caulobacter is about 3hrs. = 6 times doubling
26 = 64 -> 0.5/64= 0.008(starting O.D)
(xml)(0.902O.D)=(250ml)(.008O.D) x=2ml
Add 2ml of culture to 250ml PYE
6. Incubate for 18hrs.
7. Split 250ml of culture among five 50ml centrifuge tubes
8. Centrifuge cells at 10000XG for 7mins at 4oC
9. Remove supernatant and resuspend cells by in 50ml of ice-cold sterile, distilled water. Repeat for all tubes. DO NOT VORTEX
10. Centrifuge w/ same conditions
11. Remove supernatant and resuspend the cells in 25ml of ice-cold, sterile, distilled water.
12. Centrifuge.
13. Remove supernatant and resuspend the cells in 1/20 the original volume (50ml in each tube)
1/20 * 50ml = 2.5ml of ice-cold, sterile 10% glycerol.
14. Centrifuge
15. Remove supernatant resuspend cells in 150ul of ice-cold, sterile 10% glycerol.
16. Dispense cells into 50ul aliquot in eppendorf tubes. We ended up w/ 15 tubes total
17. Store at 80oC.
8/30
CB15 Transformation
Aim : Transforming CB15 w/ PxYFPC-2 and PvCFPC-4 plasmids
Materials :
-CB15(from Stanford)
-PxYFPC-2
-PvCFPC-4
-PYE+Kanamycin plates (x3)
-PYE+Gentamycin plates (x3)
-ice-cold sterile dH2O
-electroporation cuvettes (x3)
-sterile beads
Procedure:
1. If frozen, thaw electro competent cells on ice.
2. Place electroporation cuvettes on ice.
3. Dilution of Plasmids:
PxYFPC-2 conc. Is 293.2ng/ml in freezer.
-to dilute add 1ul of stock conc. into 100ul of sterile cold dh20. Making the final conc. 2.932ng/ul -> label this Px.
PcCFPC-4 conc. is 302.3 ng/ul in freezer
-to dilute add 1ul of stock conc. into 100ul of sterile cold dH20. Making the final conc. 3.023ng/ul -> label this Pv.
4. Pipet 100ul of electro competent CB15 cells to a electroporation cuvette.
5. Add 20ng of plasmid DNA so add 7ul of diluted plasmid DNA (PxYFPC-2)
6. Mix around using pipette
7. Electroporate at 2.5kV
8. Record time constant.
9. Take cuvette and immediately add 600ul of PYE. Mix using pipette.
10. Transfer to microcentrifuge tube. Then to a shaking incubator at 33oC / 300rpm/ for 2hrs(we only left it in there for 1hrs)
11. Repeat steps 5-11 for the second plasmid DNA (PvCFPC-4) and negative control. Instead of using plasmid DNA add 7ul of sterile dH20 for negative control.
-Time constants a. pxyfpc-2 5.0
b. pvcfpc-4 5.2
b. negative 4.8
time constant b/w 3.6-4.3 msec are good (depends on the machine)
12. Plating:
Plating scheme:
<img border="0" width="623" height="344" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image002.jpg"/>
13. For each tube of 600ul aliquot of transformed cells put 300ul in Kan plate and the other 300ul into Gent plate. Repeat this procedure 2 more times for negative control and the other transformation.
14. Incubate plates at 33oC for 3 days.
9/2 (Monday)
After 3 days of incubation, results are as follows
<img border="0" width="624" height="307" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image004.jpg"/>
-There are contaminants in the negative control.
-Plates w/ transform cells turned out pretty well. The results are as expected. Since PxYFPC-2 is kanamycin resistant and PvCFPC-4 is gentamycin resistant, transformation w/ PxYFPC-2 has growth only on PYE+KAN plate and PvCFPC-4 has growth on PYE+GENT plate.
9/3
Production of Biofilm (Draft)
Aim: To grow high density biofilms.
Materials:
-glass coverslips x6
-filter paper x6
-12 well culture plate
-pye+ amp
-preculture – prepared on 9/2.
Procedure :
1. Check O.D of preculture made on 9/2 by Rolando. 0.D = 1.250
Preculture: a colony was inoculated into a test tube w/ 5ml pye.
2. Autoclave glass coverslips (20min sterilization, 20 min dry)
3. Flame forcep and prepare 100ml PYE + AMP(20ul)
4. Cut filter paper into 2 pieces per sheet ( cut out 8 pieces)
5. Place filter paper/glass in wells.
-modified design w/ 7 wells with filter paper and 5 wells with glass.
6. Fill each wells with 2ml PYE+AMP
7. Inoculate 10 wells w/ 10ul of preculture culture and use 2 wells for negative control (one for glass and one for filter paper)
8. Incubate at 33oC at 350rpm.
Results : Images are obtained from bright field microscope under 40x after 20-24 hrs. of growth. There are no visible cells on filter paper(left) oppose to a semi uniform layer of biofilm forming on the glass surface(right)
<img border="0" width="286" height="215" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image006.jpg"/> <img border="0" width="294" height="214" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image008.jpg"/>
9/6
Inducing xylose and vanillate
Aim: To inducing transform cells w/ 0.03% xylose or .5mM vanillate on agar plates.
Materials:
-pye+gent agar plate(x2)
-pye+kan agar plate(x2)
-1% xylose
-50mM vanillate
Procedure:
Plating scheme:
<img border="0" width="344" height="286" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image010.jpg"/>
1. Calculate the amount of 1% xylose to add on agar plates
- Xylose = 0.03%
15ml(0.03g/100ml) = x (.25g/25ml)
x = .45ml = 450ul
squirt 450ul of 1% xylose one pye + kan and one pye + gent plate. Wait 15mins until they dry
2. Calculate the amount of 50mM vanillate to add on agar plates (pye +gent)
- Vanillate = 0.5mM
50mM x = (15 + x )(0.5mM) x= 151.5ul
squirt 151.1ul of 50mM vanillate one pye+ gent and one + kan plate. Wait until they dry
3. Pick a colony from CB15 + PxYFPC-2 transform plate and streak on a kan plate with xylose. Repeat for the other kan plate with vanillate.
4. Repeat step 3 for PvCFPC-4 on gent plates.
5. Incubate for 2 or 3 days at 33oC
6. Look at plates under UV lamp
Results: There is no difference in fluorescent b/w induced and non-induced plates.
9/5
Protocol for 50mM vanillate
Aim: to make 50mM vanillate from vanillic acid.
Materials:
-0.21g vanillic acid [lot b185020]
-naoh
-centrifuge tube
-.2 micron filter
-syringe
Procedure:
Calculation
X = (0.050)(0.025)(168.5) = 0.210 g vanillic acid
1. Add 0.210g of vanillic acid acid into a centrifuge tube along w/ 25ml dh20.
2. Vortex solution. Until it homogenizes
3. Titrate w/ NaOH until pH reaches 7.4
9/9
Inducing xylose or vanillate into liquid cultures.
Aim: Inducing xylose or vanillate into liquid cultures.
Materials:
-PYE + Kan
-PYE+ Gent
-50mM vanillate
-50% xylose
-glass test tubes (x4)
-50ml centrifuge tubes (x4)
Procedure:
Calculations:
2mM vanillate in 25ml PYE from 50mM stock concentration vanillate
X(50mM)= ( 10+x)(2) x= 412ul
1% xylose in 10ml PYE from 50% stock concentration xylose.
X(50mM)= ( 10+x) x= 204ul
1. Add 5ml of PYE+Kan to a test tube, then inoculate it w/ pxyfpc-2 by picking a colony from transformed plate. Make a negative
2. Repeat step 1 for pvcfpc-4. Use pye + gent instead of pye + kan
3. Incubate at 33oC 300rpm overnight.
4. Prepare 4 tubes, two w/ 10ml pye+kan and the rest w/ pye + gent.
5. For the two 10ml pye+kan inoculate each tube w/ 2.0ml of overnight preculture(pxyfpc-2). Do the same for pvcfpc-4 (2ml of preculture(pvcfpc-4) into pye+gent)
6. Wait a couple of hours until O.D reaches 0.4-0.6.
O.D 0.537 when induce
7. Inoculate one pye+kan tube w/ 416ul of 50% xylose. The second one is negative control (Noninduced)
8. Inoculate one pye +gent tube w/ 204ul of 50mM vanillate. Second one is noninduced
9. Incubate at 33oC 300rpm overnight.
10. Check if cells fluoresce under w/ U.V lamp.
Results: Induction w/ 2mM vanillate ( 2 tubes next to each other on the right) doesn’t seem to fluoresce under U.V light. On the contrary tube(far left) w/ 1% has a different color compare to its’ noninduced negatice control <img border="0" width="500" height="301" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image012.jpg"/>
9/10
1X PBS (phosphate buffer saline) 1L
Aim: To make PBS for DAPI staining
Materials:
-8g of NaCl 137mM NaCl
-0.2g of KCl [lot 123796] 2.7mM KCl
-1.44g of Na2HPO4 [lot 116747] 10mM Na2HPO4
-0.24g KH2PO4 2mM KH2PO4
-HCl
Procedure:
1. Recipe above is for 1L. We’re only making 500ml so divide everything by 2.
2. Add 4g NaCl, 0.1g KCl, 0.72g Na2HPO4, and 0.12g KH2PO4 to a bottle
3. Then add dH20 to the 500ml mark
4. Titrate with HCl until pH reaches 7.4
9/11
Stock solution of DAPI
Aim: to make stock solution of DAPI
Materials:
-10mg DAPI
-2ml dH2O
-20 eppendorf tubes.
Procedure:
1. Add 2ml dH2O into 10mg DAPI vial.
2. Vortex vial.
3. Aliquot 10ul into eppendorf tubes.
4. Store in freezer. Away from light.
9/11-9/12
Vanillate and xylose induction at different concentration.
Aim: to find a concentration at which transform cells fluoresce.
Materials:
-12 well culture plate (x3)
-CB15+PxYFPC-2 preculture (from previous experiment)
-CB15+PvCFPC-4 preculture (from previous experiment)
-CB15+PvCFPC-4 pick from a plate
-PYE+KAN
-PYE+GENT
-50% xylose
-50mM vanillate
Procedure:
9/11
1. Precultures of CB15+PxYFPC-2 and CB15+PvCFPC-4 were prepared from previous induction experiment. 5ul of preculture was added to new 5ml PYE.
2. Pick a colony from pvcfpc-4 transformed plate. Prepare a negative.
3. Incubate at 200rpm 33oC
9/12
4. Put 2ml of PYE+KAN in 6 wells on a culture plate. Repeat twice for PYE+Gent on new plates. (one gent would from preculture and the other one from a new colony picked from plate)
5. Inoculate all 2ml PYE+Kan wells w/ 250ul of pxyfpc-2 preculture.
6. For Pye+Gent inoculate one plate w/ 250ul of pvcfpc-4 preculture and the other plate w/ preculure from colony picked from plate
7. Incubate at 33oC 200rpm until O.Ds of all cultures are round 0.4-0.6. Then induce according to the following scheme. <img border="0" width="554" height="229" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image014.jpg"/>
8. Incubate at 200rpm 33oC for atleast 5 hrs.
9. Look under uv lamp for floursecent
Results.
For PxYFPC-2 there seems to be fluorescent at 0.5% xylose.
For PvCFPC-4 cells fluoresce at 5mM to 7mM
Protocol for DAPI staining
Refer to <a href="http://www.igsb.org/uploads/pdf/ProtocolDAPIStaining.pdf">http://www.igsb.org/uploads/pdf/ProtocolDAPIStaining.pdf</a>
Aim: To stain biofilms w/ DAPI
Materials:
-4ml of 4% PFA(paraformaldehyde) look at protocol
-1X PBS: look at protocol
-4ml of 0.1% Triton-X-100: look at protocol
-4ml of DAPI(stock conc. 1ug/ml): look at protocol
-fume hood
-Nutator
-biofilm coverslip
-12 well culture plate
-paper towel and aluminum foil for disposal for PFA
Procedure:
1. Make 4% PFA, 1X PBS, 0.1% Triton-X-100, and DAPI stock conc. 1ug/ml (prepare these by looking at protocols). Do all work w/ PFA in fume hood.
2. Using a new 12 well culture plate, fill a well up w/ 4ml of DH20.
3. Take a coverslip to be analyze, wash slip by transferring it to the well w/ 4ml of dH2O for 2mins w/ gentle shaking (in order to get rid of swarmer and unattached cells.)
4. Do this next step in fume hood, since we are going to work w/ PFA. Aspirate dH2O and replace medium w/ 4ml of 4% PFA. Leave coverslip in solution for 8 minutes and place plate on a nutator. Do not leave it in there for more than 10mins.
5. Aspirate 4% PFA medium. PFA is volatile so dumped liquid waste onto paper towels that should be wrapped in aluminum foil before disposal.
6. Wash cells by inoculating 4ml of 1x PBS into well for 5mins. Place plate on a nutator. Repeat this process 2 more times (wash cells w/ 1x PBS for a total of 3 times.)
7. Remove 1X PBS and add 4ml of 0.1% Triton-X-100 (to permeabilize cells). Leave coverslip in there for 10mins. Also place plate on a nutator.
8. After 10mins, repeat step 6. Wash cells 3 times w/ 1X PBS (5mins/wash.)
9. Stain cells by inoculating well w/ 4ml of DAPI for 5mins. Place plate on a nutator.
10. Wash cells w/ 1X PBS for 5 mins.
11. Coverslip is now ready to be analyzed
12. See microscopy protocol.
9/16-9/20
Production of Biofilm
Aim : To grow a high density uniform monolayer under varying parameters. We will culture the bacteria for 4 days taking data points at various time intervals. The cultures will be grown on glass cover slips, micro-filter paper, and plastic.
Materials :
C. crescentus CB15 (pre-culture)
PYE + ampicillin
Clorox
Ethanol
Milli HQ water
12 Well Tissue Culture Plate x 10
40 Glass coverslips 15mm
40 Filter Paper 0.22 micron
40 Plastic coverslips
forceps
Protocol :
1) Confirm if supply of pre-culture is available.
Preparing a pre culture: Using a pipette tip pick a colony from CB15 plate (from Stanford) dip it in 5ml PYE+amp. Incubate this at 200rpm and 33degrees C overnight.
2) Prepare glass cover slips by cleaning with w/ ethanol. There’s 10% ethanol in lab if not we can make 10% ethanol by adding 100ml of ethanol to 900ml dH20. Squirt some ethanol on coverslips and wipe them away with KimWipes.
3) Next sterilize glass pieces, PYE, dH2O, and forceps by autoclave at 120°C for 40 mins on fluid cycle
4) Sterilize plastic pieces by using ethanol and rinsing off w/ sterile dH20
See figure for layout (these steps done for each different surface type)
5) Place a cover slip in each well of tissue culture plate. We will have a total of 10 culture plates testing 3 different parameters (temps, days, and replacement of nutrients) See figure for experiment layout.
<img border="0" width="623" height="761" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image016.jpg"/>
6) Add 2mL of PYE to each well.
7) Identify negative control wells.
8) Before inoculating check OD600 of preculture.
Procedure for spectrometer: Turn on the spectrometer, there’s a switch on the back of the machine. Hit option 4 Cell Count. Zero the machine out w/ a blank (do this by filling a cuvette w/ PYE, place it in the machine, and hit auto zero.) There should be an arrow on the cuvette, turn the arrow until it faces left and place it in the spectrometer. Read numbers off the top right. It should say 600nm and 0.00. Next pipette out 200ul of culture into another cuvette, place it in the machine, and read off the top right corner or press start and it’ll pop out a number (this is the OD at 600nm).
9) Inoculate each well that is to receive culture with 10uL of pre culture (CB15)
10) Incubate at 33°C at 300rpm in incubator
To take a data point
11 to take a data point at the 24hr interval, remove the coverslip to be analyzed. Use sterile forcep to handle coverslip. Wash coverslip to remove swarmer and unattached stalked cells by preparing a petri dish filled w/ 15ml PBS (can also use a well and fill it w/ 2ml dh20). Place coverslip in PBS water for 2 min with gentle shaking.
(Rough graph for planning. Data points will include pictures of different staining methods. Look at different a protocol for staining)
28°C
|
24 hrs. nutrient replacement |
24hrs |
48hrs |
72hrs |
96hrs |
||||
|
Glass |
||||||||
|
Porous |
||||||||
|
Plastic |
||||||||
|
No Nutrient Replacement |
||||||||
|
Glass |
||||||||
|
Porous |
||||||||
|
Plastic |
33°C
|
24 hrs. nutrient replacement |
24hrs |
48hrs |
72hrs |
96hrs |
||||
|
Glass |
||||||||
|
Porous |
||||||||
|
Plastic |
||||||||
|
No Nutrient Replacement |
||||||||
|
Glass |
||||||||
|
Porous |
||||||||
|
Plastic |
PYE+AMP
13) return culture plate to incubator.
14) analyze removed cover slip under light microscopy. (see protocol)
9/23
Protocol for 0.1% Triton-X-100
Aim: To make 0.1% Triton-X-100 for DAPI staining
Materials:
-100% Triton-X-100
-250ml of sterile PBS
-sterile glass bottle for 250ml solution
Procedure:
1. Add 250ul of 100% Triton X-100 and 249.75ml of dH20 to a glass bottle
2. Storage: Store in tightly closed containter in a cool well-ventillated (area around 25C).
MSDS: <a href="https://fscimage.fishersci.com/msds/24515.htm">https://fscimage.fishersci.com/msds/24515.htm</a>
<a href="http://www.sigmaaldrich.com/etc/medialib/docs/Sigma/Product_Information_Sheet/1/t8532pis.Par.0001.File.tmp/t8532pis.pdf"> http://www.sigmaaldrich.com/etc/medialib/docs/Sigma/Product_Information_Sheet/1/t8532pis.Par.0001.File.tmp/t8532pis.pdf </a> (storage info)
8/15
Peptone Yeast Extract (PYE) Protocol
(for preparation of one Liter)
Aim : To make 1 Liter of PYE growth medium for Caulobacter
Materials :
Bacto Peptone 0.2% approx. 2g lot# 1194403
Yeast Extract 0.1% approx. 1g lot# 1307388 vat# 288620
MgSO4 1mM 0.247g EMD#MX0070-1 lot# uk09FZEMS
CaCl2 o.5mM 0.074g lot# A0141702020
1 Liter flask
milliHQ water
autoclave
Procedure :
1) Combine 2g of bacto peptone, 1g of yeast extract, 0.24624g of MgSO4, and 0.07350g of CaCl2 in a sterile 1 Liter flask.
2) Fill flask with millliHQ water to 1 Liter mark on flask.
3) Homogenize mixture by shaking/stiring.
4) Sterilize mixture using the autoclave on fluid cycle for 40 minutes.
5) Let cool, cover, and store in refrigerator.
Refer to <a href="http://labs.mcdb.lsa.umich.edu/labs/maddock/protocols/media/antibiotics.html"> http://labs.mcdb.lsa.umich.edu/labs/maddock/protocols/media/antibiotics.html </a> for Antibiotics Concentations.
4/9/93
Antibiotics
|
Antibiotic |
Stock conc (mg/ml) |
Final conc (μg/ml) |
|||||
|
E. coli (Big Red) |
E. coli LB Plates* |
Caulobacter PYE plates * |
Caulobacter liquid media |
||||
|
Ampicillin |
100 |
100 |
100 |
50 |
5 - 10 |
||
|
Chloramphenicol, in ethanol |
5 |
20 |
30 |
1 |
|||
|
Gentamycin |
2.5 |
15 |
20 |
5 |
2.5 |
||
|
Kanamycin |
50 |
30 |
50 |
20 |
5 |
||
|
Nalidixic acid, sodium salt |
10 |
15 |
20 |
20 |
- |
||
|
Oxytetracycline, in 70% EtOH |
5 |
12 |
12.5 |
2 |
1 |
||
|
Spectinomycin |
10 |
100 |
50 |
50 |
25 |
||
|
Streptomycin |
50 |
30 |
30 |
5 |
5 |
||
Rule of thumb for CB in liquid media
: Use half the conc. of antibiotic used in solid media.
Notes:
Storage: All antibiotics should be stored at 4° C, Tet should be stored at -20° C.
Tet is also light sensitive. Store stock solutions & plates in the dark.
Prep: All antibiotics should be dissolved in sterile distilled water unless otherwise indicated.
Use oxytetracycline for Caulobacter strains.
8/15
Innoculating Ecoli PxYFPC-2
Aim: Picking Ecoli from a plate to get PxYPFC-2 plasmids
Materials:
-Ecoli PxYFPC-2 plate from Stanford
-LB growth medium (made by Aaron)
-Kanamycin ( stock conc. 50mg/ml)
-Inoculating Loop
-Flame
Procedure:
1. Set up a flame
2. Sterilize the loop by heating it over the flame
3. Using the inoculating loop, pick a colony
4. Dip loop into sterile LB broth (100ml)
5. Store in shaking incubator at -37oC for 1 night
8/19 Agar Plates
Aim: To make PYE + antibiotic agar plates
Materials:
-Petri dishes
-PYE (500ml- this is enough to make 15-18 plates)
-Kanamycin/Gentamycin
-Agar (13-15 g per liter) [Lot # 2107450]
-1 Liter Erlenmeyer Flask
Procedure:
1. Add 7.5g of agar and 500mL of PYE to a 1L flask.
2. Homogenize mixture using a stirring bar.
3. Autoclave for 40 mins on fluid cycle.
4. Cool mixture to 50°C
5. add antibiotic to mixture:
6. For antibiotic concentrations refer to:
“A comprehensive set of plasmids for vanillate and xylose-inducible gene expression in Caulobacter Cresentus”
for kanamycin plates: add 250uL kanamycin
for gentamycin plates: add 250uL gentamycin
7. Pour agar into sterilized petri dishes.
8. Let stand for 15 mins to allow agar to solidify.
9. Cover with parafilm and store in refrigerator.
8/26
Ampicillin dilution
Aim: to make stock ampicillin 50mg/ml
Materials:
-Ampicillin [Lot # 29371]
-BD syringe 0.2micron filtuer
-50ml centrifuge tube
-eppendorf tubes
Procedure:
1. Weight out 0.5 g of Ampicillin
2. Place in 50ml centrifuge tube along w/ 10ml of dH2O.
3. Vortex the solution
4. Using BD syringe filter out 10ml of diluted antibiotic into eppendorf tubes.
5. Fill 20 eppendorf tubes w/ aliquot of 500ul.
8/28
Growth Curve
Aim : to create a standard caulobacter growth curve
Materials:
-PYE+AMP
-50ml centrifuge tubes x6
-CB15
Procedure:
We’re going to test 2 different temps. 28*c and 33* c
For 33 C (Rolando)
1. Label 1 centrifuge tube “negative” and the other two c tubes. “33 1” and “33 1b”
2. Make 80ml of pye + amp. Look on page _______ for PYE protocol and page______ for AMP dilution
Final conc of amp = 10ug/ml
Add 80ml of pye into a flask w/ 16ul of ampicillin( stock conc. 50mg/ml)
3. Fill each tube w/ 4ml PYE
4. For negative control tube dip a sterile pipette tip in the tube. Discard tip.
5. For the other 2 tubes pick a colony from CB15 plate w/ a pipette tip. Pick two colonies one for each.
6. Store in incubator at 33C at 300rpm
7. Make a table and later transfer in unto a plot of O.D600 versus time graph. ( should have a total of 3 tables and 3 graphs)
8. Record an OD reading every 2 hrs.
Negative control 33C 33C b
|
Time |
OD600 |
Time |
OD600 |
Time |
OD600 |
For 33 C (Linh)
1. Label 1 centrifuge tube “negative” and the other two c tubes. “33 2” and “33 2b”
2. Make 80ml of pye + amp. Look on page _______ for PYE protocol and page______ for AMP dilution
Final conc of amp = 10ug/ml
Add 80ml of pye into a flask w/ 16ul of ampicillin( stock conc. 50mg/ml)
3. Fill each tube w/ 4ml PYE
4. For negative control tube dip a sterile pipette tip in the tube. Discard tip.
5. For the other 2 tubes pick a colony from CB15 plate w/ a pipette tip. Pick two colonies one for each.
6. Store in incubator at 33C at 300rpm
7. Make a table and later transfer in unto a plot of O.D600 versus time graph. ( should have a total of 3 tables and 3 graphs)
8. Record an OD reading every 2 hrs.
8/26
Inoculation of CB15
Aim: making liquid culture of CB15
Materials:
-CB15(from Stanford)
-Antibiotic
-PYE
Procedure:
1. Add 50ul of Ampicillin (50mg/ml) into 250ml of PYE
Refer to for final concentration of Ampicillin(10ug/ml):
“Dynamics and Control of Biofilms of the Oligotrophic Bacterium Caulobacter Crecentus.”
2. Pipet 20ml of pye + amp into centrifuge tube. Do this twice. Label one “Negative” and the other “CB15”
3. Pick a colony from CB15 plate and eject tip in tube
4. For negative control, drop a blank pipet tip
5. Incubate at 28°C and 180 rpm
Results:
After 1 night PYE medium for negative control turned cloudy. Experiment has been contaminated.
8/29
Making Glycerol Stocks
Aim: for long term storage of CB15
Materials:
-glycerol 60% (filtered)
-cell culture cb15
-eppendorf tubes.
Procedure:
1. Prepare a preculuture: Pick a CB15 colony and inoculate it in 5ml of PYE. When culture reaches stationary phase the O.D600 should be around 1.3-1.6
2. Caulobacters are store in 10% glycerol
Refer to: “Caulobacter S-Layer Secretion Kit Manual”
3. Calculate 10% glycerol.
(x amount of 10% glycerol )(60)= (900ul of cells + x)(10)
This comes out to be 180ul of 60% glycerol per 900ul of cb15
4. In an eppendorf tube combine 180ul of 60% glycerol and 900ul of cb15 culture.
5. Shake tube.
6. Label and store tubes at -80C
8/29-8/30
Preparing Electrocompetent CB15
Aim : making electrocompetent CB15
Materials:
-PYE
-eppendorf tubes
-centrifuge tubes(50ml)
-1L falsk
-ice-cold, sterile, distilled water
-ice-cold, sterile 10% glycerol in distilled water.
Procedure : will yield enough electro competent cells for 15 transformation.
1. Diluting 60% stock conc. Glycerol into 10%
Add 80ml of distilled water and 16ml of 60% filtered glycerol from fridge into a flask
2. Autoclave 1000ml of distilled water, 250ml PYE flask (x2), 96ml of 10% glycerol - fluid cycle 40mins
3. Inoculate a CB15 colony into 5ml PYE medium. Grow at 30oC w/ shaking until the O. D is 0.5-1.0 ( should take about 1 night)
4. Record the O.D of the culture above. 0.902
5. Dilution calculation: calculate the amount of culture you would add to 250ml of fresh PYE to achieve an O.D of 0.4-0.6 in 18hrs.
-doubling time of Caulobacter is about 3hrs. = 6 times doubling
26 = 64 -> 0.5/64= 0.008(starting O.D)
(xml)(0.902O.D)=(250ml)(.008O.D) x=2ml
Add 2ml of culture to 250ml PYE
6. Incubate for 18hrs.
7. Split 250ml of culture among five 50ml centrifuge tubes
8. Centrifuge cells at 10000XG for 7mins at 4oC
9. Remove supernatant and resuspend cells by in 50ml of ice-cold sterile, distilled water. Repeat for all tubes. DO NOT VORTEX
10. Centrifuge w/ same conditions
11. Remove supernatant and resuspend the cells in 25ml of ice-cold, sterile, distilled water.
12. Centrifuge.
13. Remove supernatant and resuspend the cells in 1/20 the original volume (50ml in each tube)
1/20 * 50ml = 2.5ml of ice-cold, sterile 10% glycerol.
14. Centrifuge
15. Remove supernatant resuspend cells in 150ul of ice-cold, sterile 10% glycerol.
16. Dispense cells into 50ul aliquot in eppendorf tubes. We ended up w/ 15 tubes total
17. Store at 80oC.
8/30
CB15 Transformation
Aim : Transforming CB15 w/ PxYFPC-2 and PvCFPC-4 plasmids
Materials :
-CB15(from Stanford)
-PxYFPC-2
-PvCFPC-4
-PYE+Kanamycin plates (x3)
-PYE+Gentamycin plates (x3)
-ice-cold sterile dH2O
-electroporation cuvettes (x3)
-sterile beads
Procedure:
1. If frozen, thaw electro competent cells on ice.
2. Place electroporation cuvettes on ice.
3. Dilution of Plasmids:
PxYFPC-2 conc. Is 293.2ng/ml in freezer.
-to dilute add 1ul of stock conc. into 100ul of sterile cold dh20. Making the final conc. 2.932ng/ul -> label this Px.
PcCFPC-4 conc. is 302.3 ng/ul in freezer
-to dilute add 1ul of stock conc. into 100ul of sterile cold dH20. Making the final conc. 3.023ng/ul -> label this Pv.
4. Pipet 100ul of electro competent CB15 cells to a electroporation cuvette.
5. Add 20ng of plasmid DNA so add 7ul of diluted plasmid DNA (PxYFPC-2)
6. Mix around using pipette
7. Electroporate at 2.5kV
8. Record time constant.
9. Take cuvette and immediately add 600ul of PYE. Mix using pipette.
10. Transfer to microcentrifuge tube. Then to a shaking incubator at 33oC / 300rpm/ for 2hrs(we only left it in there for 1hrs)
11. Repeat steps 5-11 for the second plasmid DNA (PvCFPC-4) and negative control. Instead of using plasmid DNA add 7ul of sterile dH20 for negative control.
-Time constants a. pxyfpc-2 5.0
b. pvcfpc-4 5.2
b. negative 4.8
time constant b/w 3.6-4.3 msec are good (depends on the machine)
12. Plating:
Plating scheme:
<img border="0" width="623" height="344" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image002.jpg"/>
13. For each tube of 600ul aliquot of transformed cells put 300ul in Kan plate and the other 300ul into Gent plate. Repeat this procedure 2 more times for negative control and the other transformation.
14. Incubate plates at 33oC for 3 days.
9/2 (Monday)
After 3 days of incubation, results are as follows
<img border="0" width="624" height="307" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image004.jpg"/>
-There are contaminants in the negative control.
-Plates w/ transform cells turned out pretty well. The results are as expected. Since PxYFPC-2 is kanamycin resistant and PvCFPC-4 is gentamycin resistant, transformation w/ PxYFPC-2 has growth only on PYE+KAN plate and PvCFPC-4 has growth on PYE+GENT plate.
9/3
Production of Biofilm (Draft)
Aim: To grow high density biofilms.
Materials:
-glass coverslips x6
-filter paper x6
-12 well culture plate
-pye+ amp
-preculture – prepared on 9/2.
Procedure :
1. Check O.D of preculture made on 9/2 by Rolando. 0.D = 1.250
Preculture: a colony was inoculated into a test tube w/ 5ml pye.
2. Autoclave glass coverslips (20min sterilization, 20 min dry)
3. Flame forcep and prepare 100ml PYE + AMP(20ul)
4. Cut filter paper into 2 pieces per sheet ( cut out 8 pieces)
5. Place filter paper/glass in wells.
-modified design w/ 7 wells with filter paper and 5 wells with glass.
6. Fill each wells with 2ml PYE+AMP
7. Inoculate 10 wells w/ 10ul of preculture culture and use 2 wells for negative control (one for glass and one for filter paper)
8. Incubate at 33oC at 350rpm.
Results : Images are obtained from bright field microscope under 40x after 20-24 hrs. of growth. There are no visible cells on filter paper(left) oppose to a semi uniform layer of biofilm forming on the glass surface(right)
<img border="0" width="286" height="215" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image006.jpg"/> <img border="0" width="294" height="214" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image008.jpg"/>
9/6
Inducing xylose and vanillate
Aim: To inducing transform cells w/ 0.03% xylose or .5mM vanillate on agar plates.
Materials:
-pye+gent agar plate(x2)
-pye+kan agar plate(x2)
-1% xylose
-50mM vanillate
Procedure:
Plating scheme:
<img border="0" width="344" height="286" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image010.jpg"/>
1. Calculate the amount of 1% xylose to add on agar plates
- Xylose = 0.03%
15ml(0.03g/100ml) = x (.25g/25ml)
x = .45ml = 450ul
squirt 450ul of 1% xylose one pye + kan and one pye + gent plate. Wait 15mins until they dry
2. Calculate the amount of 50mM vanillate to add on agar plates (pye +gent)
- Vanillate = 0.5mM
50mM x = (15 + x )(0.5mM) x= 151.5ul
squirt 151.1ul of 50mM vanillate one pye+ gent and one + kan plate. Wait until they dry
3. Pick a colony from CB15 + PxYFPC-2 transform plate and streak on a kan plate with xylose. Repeat for the other kan plate with vanillate.
4. Repeat step 3 for PvCFPC-4 on gent plates.
5. Incubate for 2 or 3 days at 33oC
6. Look at plates under UV lamp
Results: There is no difference in fluorescent b/w induced and non-induced plates.
9/5
Protocol for 50mM vanillate
Aim: to make 50mM vanillate from vanillic acid.
Materials:
-0.21g vanillic acid [lot b185020]
-naoh
-centrifuge tube
-.2 micron filter
-syringe
Procedure:
Calculation
X = (0.050)(0.025)(168.5) = 0.210 g vanillic acid
1. Add 0.210g of vanillic acid acid into a centrifuge tube along w/ 25ml dh20.
2. Vortex solution. Until it homogenizes
3. Titrate w/ NaOH until pH reaches 7.4
9/9
Inducing xylose or vanillate into liquid cultures.
Aim: Inducing xylose or vanillate into liquid cultures.
Materials:
-PYE + Kan
-PYE+ Gent
-50mM vanillate
-50% xylose
-glass test tubes (x4)
-50ml centrifuge tubes (x4)
Procedure:
Calculations:
2mM vanillate in 25ml PYE from 50mM stock concentration vanillate
X(50mM)= ( 10+x)(2) x= 412ul
1% xylose in 10ml PYE from 50% stock concentration xylose.
X(50mM)= ( 10+x) x= 204ul
1. Add 5ml of PYE+Kan to a test tube, then inoculate it w/ pxyfpc-2 by picking a colony from transformed plate. Make a negative
2. Repeat step 1 for pvcfpc-4. Use pye + gent instead of pye + kan
3. Incubate at 33oC 300rpm overnight.
4. Prepare 4 tubes, two w/ 10ml pye+kan and the rest w/ pye + gent.
5. For the two 10ml pye+kan inoculate each tube w/ 2.0ml of overnight preculture(pxyfpc-2). Do the same for pvcfpc-4 (2ml of preculture(pvcfpc-4) into pye+gent)
6. Wait a couple of hours until O.D reaches 0.4-0.6.
O.D 0.537 when induce
7. Inoculate one pye+kan tube w/ 416ul of 50% xylose. The second one is negative control (Noninduced)
8. Inoculate one pye +gent tube w/ 204ul of 50mM vanillate. Second one is noninduced
9. Incubate at 33oC 300rpm overnight.
10. Check if cells fluoresce under w/ U.V lamp.
Results: Induction w/ 2mM vanillate ( 2 tubes next to each other on the right) doesn’t seem to fluoresce under U.V light. On the contrary tube(far left) w/ 1% has a different color compare to its’ noninduced negatice control <img border="0" width="500" height="301" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image012.jpg"/>
9/10
1X PBS (phosphate buffer saline) 1L
Aim: To make PBS for DAPI staining
Materials:
-8g of NaCl 137mM NaCl
-0.2g of KCl [lot 123796] 2.7mM KCl
-1.44g of Na2HPO4 [lot 116747] 10mM Na2HPO4
-0.24g KH2PO4 2mM KH2PO4
-HCl
Procedure:
1. Recipe above is for 1L. We’re only making 500ml so divide everything by 2.
2. Add 4g NaCl, 0.1g KCl, 0.72g Na2HPO4, and 0.12g KH2PO4 to a bottle
3. Then add dH20 to the 500ml mark
4. Titrate with HCl until pH reaches 7.4
9/11
Stock solution of DAPI
Aim: to make stock solution of DAPI
Materials:
-10mg DAPI
-2ml dH2O
-20 eppendorf tubes.
Procedure:
1. Add 2ml dH2O into 10mg DAPI vial.
2. Vortex vial.
3. Aliquot 10ul into eppendorf tubes.
4. Store in freezer. Away from light.
9/11-9/12
Vanillate and xylose induction at different concentration.
Aim: to find a concentration at which transform cells fluoresce.
Materials:
-12 well culture plate (x3)
-CB15+PxYFPC-2 preculture (from previous experiment)
-CB15+PvCFPC-4 preculture (from previous experiment)
-CB15+PvCFPC-4 pick from a plate
-PYE+KAN
-PYE+GENT
-50% xylose
-50mM vanillate
Procedure:
9/11
1. Precultures of CB15+PxYFPC-2 and CB15+PvCFPC-4 were prepared from previous induction experiment. 5ul of preculture was added to new 5ml PYE.
2. Pick a colony from pvcfpc-4 transformed plate. Prepare a negative.
3. Incubate at 200rpm 33oC
9/12
4. Put 2ml of PYE+KAN in 6 wells on a culture plate. Repeat twice for PYE+Gent on new plates. (one gent would from preculture and the other one from a new colony picked from plate)
5. Inoculate all 2ml PYE+Kan wells w/ 250ul of pxyfpc-2 preculture.
6. For Pye+Gent inoculate one plate w/ 250ul of pvcfpc-4 preculture and the other plate w/ preculure from colony picked from plate
7. Incubate at 33oC 200rpm until O.Ds of all cultures are round 0.4-0.6. Then induce according to the following scheme. <img border="0" width="554" height="229" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image014.jpg"/>
8. Incubate at 200rpm 33oC for atleast 5 hrs.
9. Look under uv lamp for floursecent
Results.
For PxYFPC-2 there seems to be fluorescent at 0.5% xylose.
For PvCFPC-4 cells fluoresce at 5mM to 7mM
Protocol for DAPI staining
Refer to <a href="http://www.igsb.org/uploads/pdf/ProtocolDAPIStaining.pdf">http://www.igsb.org/uploads/pdf/ProtocolDAPIStaining.pdf</a>
Aim: To stain biofilms w/ DAPI
Materials:
-4ml of 4% PFA(paraformaldehyde) look at protocol
-1X PBS: look at protocol
-4ml of 0.1% Triton-X-100: look at protocol
-4ml of DAPI(stock conc. 1ug/ml): look at protocol
-fume hood
-Nutator
-biofilm coverslip
-12 well culture plate
-paper towel and aluminum foil for disposal for PFA
Procedure:
1. Make 4% PFA, 1X PBS, 0.1% Triton-X-100, and DAPI stock conc. 1ug/ml (prepare these by looking at protocols). Do all work w/ PFA in fume hood.
2. Using a new 12 well culture plate, fill a well up w/ 4ml of DH20.
3. Take a coverslip to be analyze, wash slip by transferring it to the well w/ 4ml of dH2O for 2mins w/ gentle shaking (in order to get rid of swarmer and unattached cells.)
4. Do this next step in fume hood, since we are going to work w/ PFA. Aspirate dH2O and replace medium w/ 4ml of 4% PFA. Leave coverslip in solution for 8 minutes and place plate on a nutator. Do not leave it in there for more than 10mins.
5. Aspirate 4% PFA medium. PFA is volatile so dumped liquid waste onto paper towels that should be wrapped in aluminum foil before disposal.
6. Wash cells by inoculating 4ml of 1x PBS into well for 5mins. Place plate on a nutator. Repeat this process 2 more times (wash cells w/ 1x PBS for a total of 3 times.)
7. Remove 1X PBS and add 4ml of 0.1% Triton-X-100 (to permeabilize cells). Leave coverslip in there for 10mins. Also place plate on a nutator.
8. After 10mins, repeat step 6. Wash cells 3 times w/ 1X PBS (5mins/wash.)
9. Stain cells by inoculating well w/ 4ml of DAPI for 5mins. Place plate on a nutator.
10. Wash cells w/ 1X PBS for 5 mins.
11. Coverslip is now ready to be analyzed
12. See microscopy protocol.
9/16-9/20
Production of Biofilm
Aim : To grow a high density uniform monolayer under varying parameters. We will culture the bacteria for 4 days taking data points at various time intervals. The cultures will be grown on glass cover slips, micro-filter paper, and plastic.
Materials :
C. crescentus CB15 (pre-culture)
PYE + ampicillin
Clorox
Ethanol
Milli HQ water
12 Well Tissue Culture Plate x 10
40 Glass coverslips 15mm
40 Filter Paper 0.22 micron
40 Plastic coverslips
forceps
Protocol :
1) Confirm if supply of pre-culture is available.
Preparing a pre culture: Using a pipette tip pick a colony from CB15 plate (from Stanford) dip it in 5ml PYE+amp. Incubate this at 200rpm and 33degrees C overnight.
2) Prepare glass cover slips by cleaning with w/ ethanol. There’s 10% ethanol in lab if not we can make 10% ethanol by adding 100ml of ethanol to 900ml dH20. Squirt some ethanol on coverslips and wipe them away with KimWipes.
3) Next sterilize glass pieces, PYE, dH2O, and forceps by autoclave at 120°C for 40 mins on fluid cycle
4) Sterilize plastic pieces by using ethanol and rinsing off w/ sterile dH20
See figure for layout (these steps done for each different surface type)
5) Place a cover slip in each well of tissue culture plate. We will have a total of 10 culture plates testing 3 different parameters (temps, days, and replacement of nutrients) See figure for experiment layout.
<img border="0" width="623" height="761" src="file:///C:/Users/Owner/AppData/Local/Temp/msohtmlclip1/01/clip_image016.jpg"/>
6) Add 2mL of PYE to each well.
7) Identify negative control wells.
8) Before inoculating check OD600 of preculture.
Procedure for spectrometer: Turn on the spectrometer, there’s a switch on the back of the machine. Hit option 4 Cell Count. Zero the machine out w/ a blank (do this by filling a cuvette w/ PYE, place it in the machine, and hit auto zero.) There should be an arrow on the cuvette, turn the arrow until it faces left and place it in the spectrometer. Read numbers off the top right. It should say 600nm and 0.00. Next pipette out 200ul of culture into another cuvette, place it in the machine, and read off the top right corner or press start and it’ll pop out a number (this is the OD at 600nm).
9) Inoculate each well that is to receive culture with 10uL of pre culture (CB15)
10) Incubate at 33°C at 300rpm in incubator
To take a data point
11 to take a data point at the 24hr interval, remove the coverslip to be analyzed. Use sterile forcep to handle coverslip. Wash coverslip to remove swarmer and unattached stalked cells by preparing a petri dish filled w/ 15ml PBS (can also use a well and fill it w/ 2ml dh20). Place coverslip in PBS water for 2 min with gentle shaking.
(Rough graph for planning. Data points will include pictures of different staining methods. Look at different a protocol for staining)
28°C
|
24 hrs. nutrient replacement |
24hrs |
48hrs |
72hrs |
96hrs |
||||
|
Glass |
||||||||
|
Porous |
||||||||
|
Plastic |
||||||||
|
No Nutrient Replacement |
||||||||
|
Glass |
||||||||
|
Porous |
||||||||
|
Plastic |
33°C
|
24 hrs. nutrient replacement |
24hrs |
48hrs |
72hrs |
96hrs |
||||
|
Glass |
||||||||
|
Porous |
||||||||
|
Plastic |
||||||||
|
No Nutrient Replacement |
||||||||
|
Glass |
||||||||
|
Porous |
||||||||
|
Plastic |
PYE+AMP
13) return culture plate to incubator.
14) analyze removed cover slip under light microscopy. (see protocol)
9/23
Protocol for 0.1% Triton-X-100
Aim: To make 0.1% Triton-X-100 for DAPI staining
Materials:
-100% Triton-X-100
-250ml of sterile PBS
-sterile glass bottle for 250ml solution
Procedure:
1. Add 250ul of 100% Triton X-100 and 249.75ml of dH20 to a glass bottle
2. Storage: Store in tightly closed containter in a cool well-ventillated (area around 25C).
MSDS: <a href="https://fscimage.fishersci.com/msds/24515.htm">https://fscimage.fishersci.com/msds/24515.htm</a>
<a href="http://www.sigmaaldrich.com/etc/medialib/docs/Sigma/Product_Information_Sheet/1/t8532pis.Par.0001.File.tmp/t8532pis.pdf"> http://www.sigmaaldrich.com/etc/medialib/docs/Sigma/Product_Information_Sheet/1/t8532pis.Par.0001.File.tmp/t8532pis.pdf </a> (storage info)
 "
"