Team:Exeter/Results
From 2013.igem.org
Fentwistle (Talk | contribs) |
Fentwistle (Talk | contribs) |
||
| Line 42: | Line 42: | ||
| + | Our output brick for the Red Light Module includes the <i>ompF</i> promoter which is specific to the protein [http://en.wikipedia.org/wiki/Porin_response:_osmoregulation_in_E.coli OmpR]. This is followed by an RBS and a restriction site for <i>BglII</i> and the coding region for a [http://parts.igem.org/Part:BBa_E0040 GFP]. A second restriction site for <i>BamHI</i> follows, then a series of STOP codons to act as a terminator. The whole sequence is flanked by the iGEM prefix and suffix allowing the BioBrick to be used in 3A Assembly. | ||
| + | We designed this BioBrick to be built by IDT in their gBlock format. | ||
The two ends of GFP have been ligated together to form a complete, working protein in the successful Gibson assembly of our output bricks. GFP is constitutively expressed within DH5α cells as endogenous ENVZ phosporylates endogenous OmpR as a component of osmolarity sensing. Endogenous OmpR then binds to OmpF. Thus our output brick is working as expected. In order to test whether the output brick can be implemented into the Red light pathway and ENVZ- strain should be used to avoid false positives. | The two ends of GFP have been ligated together to form a complete, working protein in the successful Gibson assembly of our output bricks. GFP is constitutively expressed within DH5α cells as endogenous ENVZ phosporylates endogenous OmpR as a component of osmolarity sensing. Endogenous OmpR then binds to OmpF. Thus our output brick is working as expected. In order to test whether the output brick can be implemented into the Red light pathway and ENVZ- strain should be used to avoid false positives. | ||
Revision as of 08:40, 1 October 2013
Green Light Module
We have had marked success with our Green Light Module. Not only is the magenta pigment bright and evenly synthesised, but we have also managed to show magenta synthesis is inhibited by exposure to white light. We are currently working on showing that green light specifically switches off magenta production.
Cyan pigment
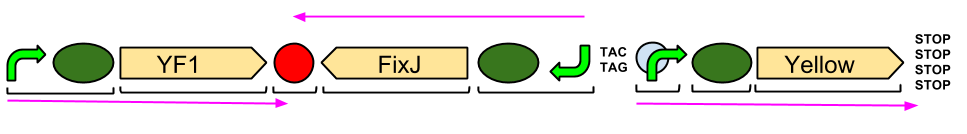
Red Light Module output Brick with BglII and BamHI restriction sites
Our output brick for the Red Light Module includes the ompF promoter which is specific to the protein [http://en.wikipedia.org/wiki/Porin_response:_osmoregulation_in_E.coli OmpR]. This is followed by an RBS and a restriction site for BglII and the coding region for a [http://parts.igem.org/Part:BBa_E0040 GFP]. A second restriction site for BamHI follows, then a series of STOP codons to act as a terminator. The whole sequence is flanked by the iGEM prefix and suffix allowing the BioBrick to be used in 3A Assembly.
We designed this BioBrick to be built by IDT in their gBlock format.
The two ends of GFP have been ligated together to form a complete, working protein in the successful Gibson assembly of our output bricks. GFP is constitutively expressed within DH5α cells as endogenous ENVZ phosporylates endogenous OmpR as a component of osmolarity sensing. Endogenous OmpR then binds to OmpF. Thus our output brick is working as expected. In order to test whether the output brick can be implemented into the Red light pathway and ENVZ- strain should be used to avoid false positives.
Planned "Post-Freeze" Work
Blue Light Module
Our Blue Light Module, which is being built by DNA2.0, will unfortunately not arrive in the labs until after the Wiki-freeze. However, we will be bringing plenty of pictures of it working (hopefully!) to the Jamboree. Considering it has been constructed in a very similar manner to the Green Light Module, which is functioning in the lab, we are reasonably hopefully that we can have the Blue Light Module functioning in the lab soon after its arrival.
Red Light Module
(Cyan working in EnvZ deficient cells)
 "
"