Team:Leeds/Project
From 2013.igem.org
(Difference between revisions)
| Line 80: | Line 80: | ||
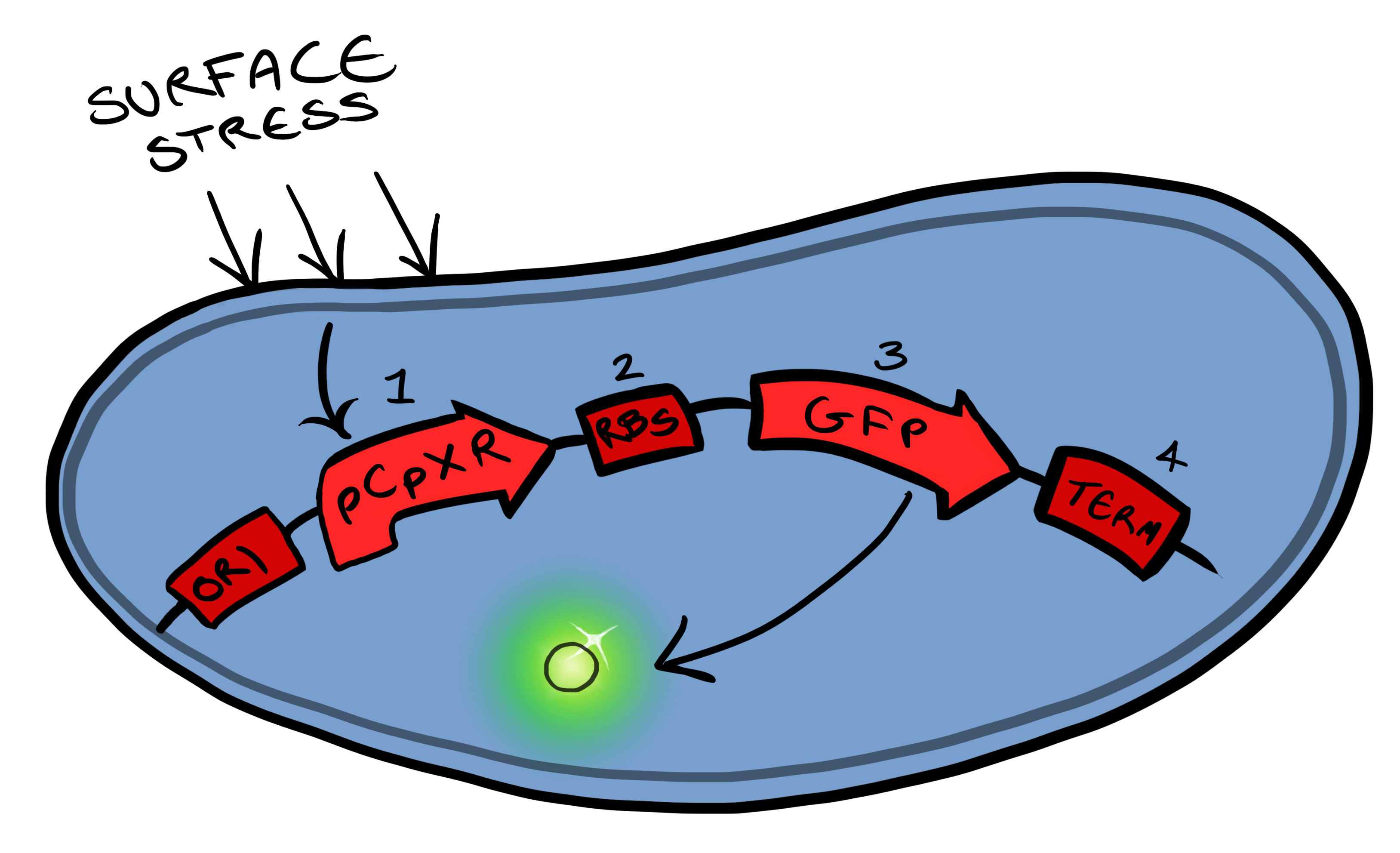
BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator | BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator | ||
<br style="clear:both"/> | <br style="clear:both"/> | ||
| - | === | + | ===Si4 Binding Peptide=== |
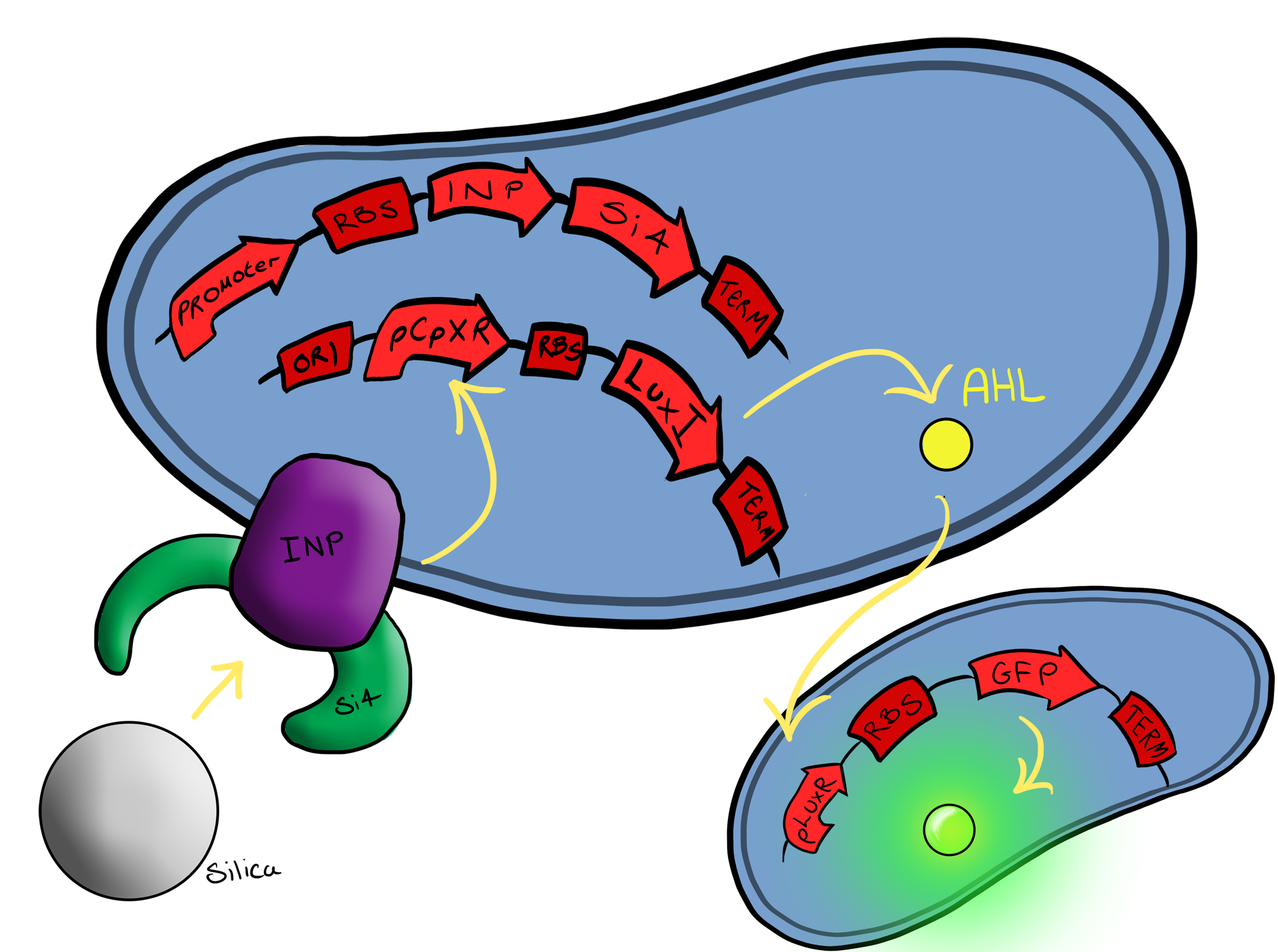
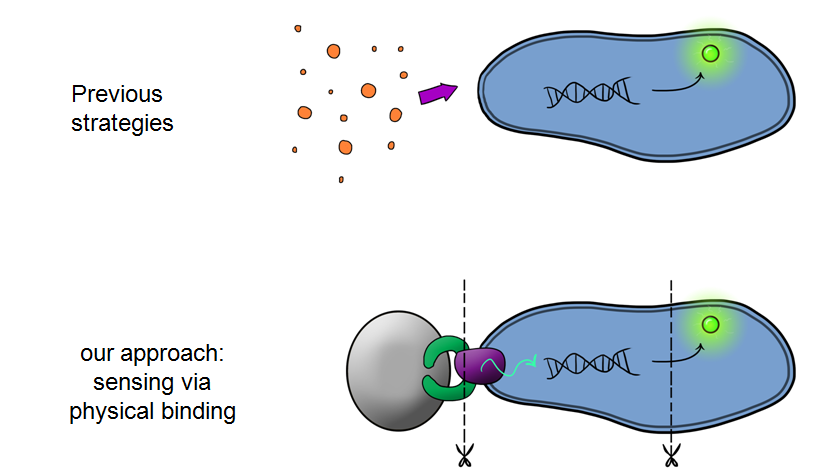
| - | + | Simultaneously, we will be developing Device 2 that is suited for physical attachment and detection of particles. This will work using Ice Nucleation Protein (INP) to display a oligo-peptide of our choice on the outer-membrane of our ''E. coli'', initially this will be a peptide capable of binding silica beads allowing us to create a model system of pathogen detection. INP is a transmembrane protein that expresses any sequence that is placed on the C-terminus of its gene on the outer surface of the cell. Check out our Lab results page to see the characteristaion results for the Si4 binding domain. | |
| - | Simultaneously, we will be developing | + | |
<br style="clear:both"/> | <br style="clear:both"/> | ||
| - | + | ||
<br style="clear:both"/> | <br style="clear:both"/> | ||
| - | The Biobricks we used for this | + | The Biobricks we used for this Device are: |
BBa_K081012 for the Green fluorescent protein generator which contain a ribosome binding site and terminator. | BBa_K081012 for the Green fluorescent protein generator which contain a ribosome binding site and terminator. | ||
BBa_K523008 is the Ice nucleation protein any sequence added to the C-terminus of the INP will be transported to cell membrane and be displayed on the surface of the cell. | BBa_K523008 is the Ice nucleation protein any sequence added to the C-terminus of the INP will be transported to cell membrane and be displayed on the surface of the cell. | ||
BBaa_B0015, which is a double terminator, | BBaa_B0015, which is a double terminator, | ||
BBa_B0034 a strong ribosome binding site and finally | BBa_B0034 a strong ribosome binding site and finally | ||
| - | + | ||
<br> | <br> | ||
==Phase II== | ==Phase II== | ||
| - | Phase II takes the products of Phase I to the next level, and begins to look at integration of the | + | Phase II takes the products of Phase I to the next level, and begins to look at integration of the Si4 binding peptide and Device 1 to give a new device; Device 2. |
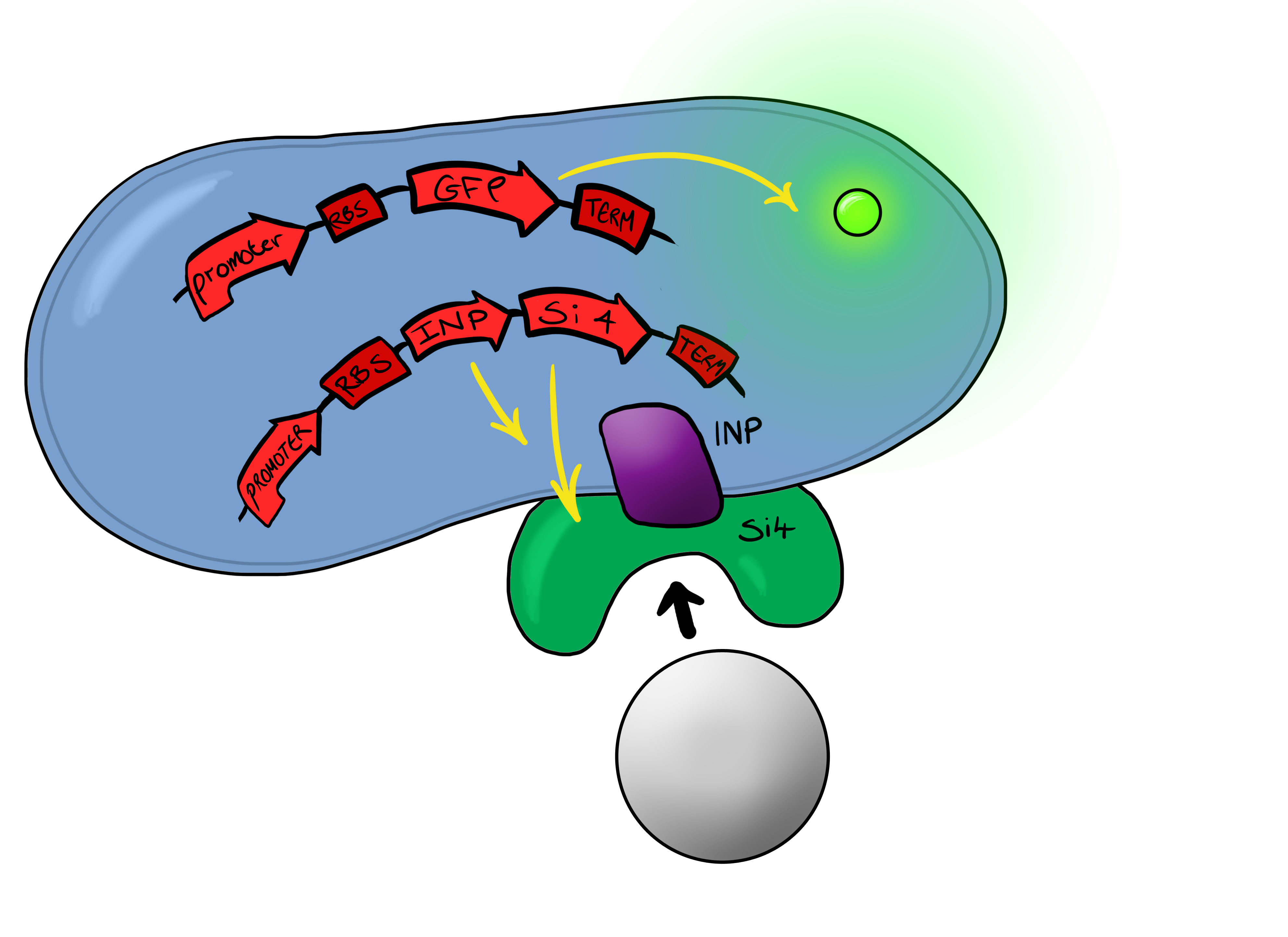
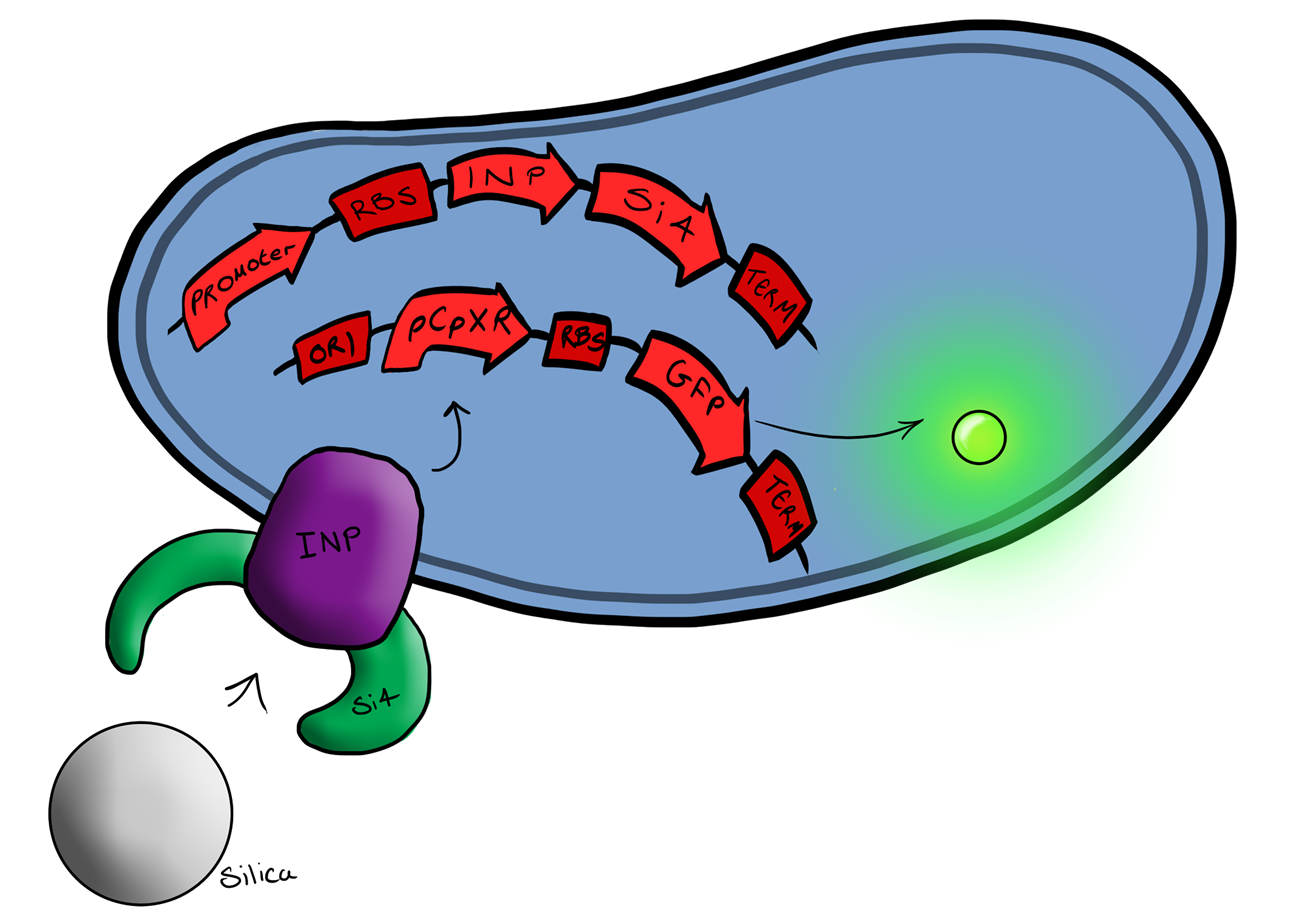
| - | ===Device | + | ===Device 2=== |
[[File:Biosystem3.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] | [[File:Biosystem3.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] | ||
| - | Device | + | Device 2 brings together BS1 and BS2 in one plasmid; the genes that code for the INP and the Si4 binding domain are now located on the same plasmid as the CpxP promoter and the GFP generator. The effect of this is that when the silca bead binds to the Si4 binding domain connected to the INP protein and causes surface stress the CpxP promoter will be induced. This will cause GFP to be produced and this will act as a visible signal that the 'pathogen' has been detected. |
<br style="clear:both"/> | <br style="clear:both"/> | ||
| - | This is the final MircoBeagle device and has the potential to be made into many different pathogen detectors. This can easily be done by swapping out the gene coding for the binding moiety next to the INP gene. Once Biosystem | + | This is the final MircoBeagle device and has the potential to be made into many different pathogen detectors. This can easily be done by swapping out the gene coding for the binding moiety next to the INP gene. Once Biosystem 2 has been suitably characterised and all bugs fixed, the next step will be to swap the silca binding domain for a binding domain that binds to a particular pathogen. |
<br style="clear:both"/> | <br style="clear:both"/> | ||
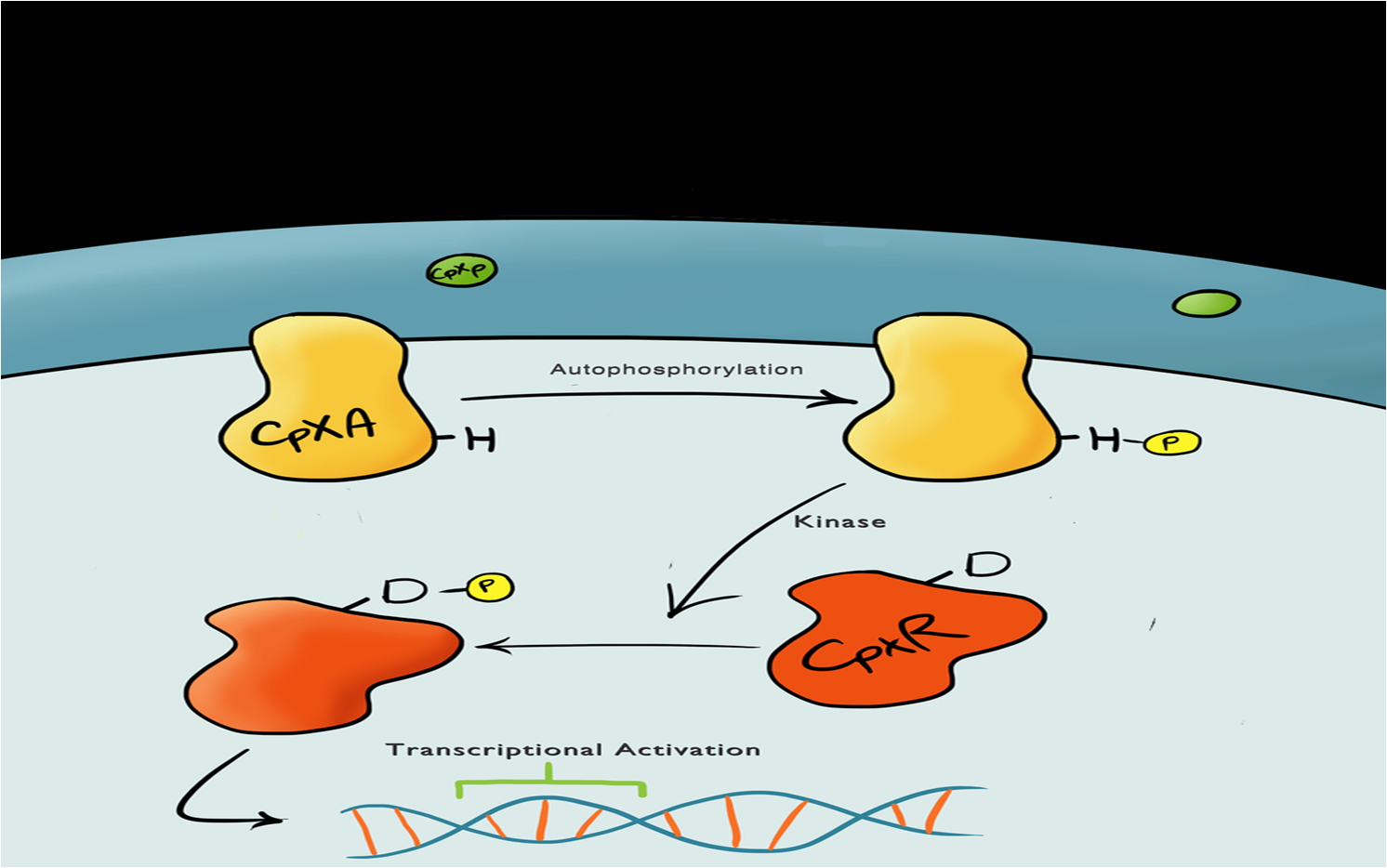
==The Cpx Pathway== | ==The Cpx Pathway== | ||
Revision as of 09:37, 1 October 2013
 "
"