Team:Braunschweig/Results
From 2013.igem.org
| Line 35: | Line 35: | ||
In our project we were able to assemble two functional devices (BBa_K1073034 and BBa_K1073035) using standard Biobricks from the registry.<br> | In our project we were able to assemble two functional devices (BBa_K1073034 and BBa_K1073035) using standard Biobricks from the registry.<br> | ||
To establish a stable microbial consortium our engineered constructs consist of different operation units. We demonstrated the functionality of each unit by individual experiments:<br> | To establish a stable microbial consortium our engineered constructs consist of different operation units. We demonstrated the functionality of each unit by individual experiments:<br> | ||
| + | |||
| + | <img alt="grey line" src="https://static.igem.org/mediawiki/2013/4/4c/Braunschweig_grey_line.png" width="850" height="1" vspace="20"/> | ||
<p style="font-size:14px; font-weight:bold; text-decoration:none; border:none; color:#be1e3c">1. Reporter-chromoproteins were successfully integrated into the final constructs</p> | <p style="font-size:14px; font-weight:bold; text-decoration:none; border:none; color:#be1e3c">1. Reporter-chromoproteins were successfully integrated into the final constructs</p> | ||
| - | The included reporter-chromoproteins were visible to the naked eye in less than 24 h during incubation on agar plates or in liquid culture (fig. 1). The use of chromoproteins as reporter facilitated the distinction of the different bacterial strains we intended to cultivate in co-culture. Featured chromoproteins were amilGFP, eforRed and aeBlue which developed a yellowish, bright pink and dark blue color.<br><br> | + | <p>The included reporter-chromoproteins were visible to the naked eye in less than 24 h during incubation on agar plates or in liquid culture (fig. 1). The use of chromoproteins as reporter facilitated the distinction of the different bacterial strains we intended to cultivate in co-culture. Featured chromoproteins were amilGFP, eforRed and aeBlue which developed a yellowish, bright pink and dark blue color.<br><br> |
<dl class="imgCenter"> | <dl class="imgCenter"> | ||
| - | <dt><img alt="Chromoproteins" src="https://static.igem.org/mediawiki/parts/thumb/c/c1/Expressed_chromoproteins.png/450px-Expressed_chromoproteins.png" width=" | + | <dt><img alt="Chromoproteins" src="https://static.igem.org/mediawiki/parts/thumb/c/c1/Expressed_chromoproteins.png/450px-Expressed_chromoproteins.png" width="300" /> |
</dt> | </dt> | ||
<dd>Fig. 1: Successful integration of reporter-chromoproteins into our constructs.<br> Liquid overnight cultures expressing the chromoproteins amilGFP (left), eforRed (middle), aeBlue (right).</dd></dl> <br> | <dd>Fig. 1: Successful integration of reporter-chromoproteins into our constructs.<br> Liquid overnight cultures expressing the chromoproteins amilGFP (left), eforRed (middle), aeBlue (right).</dd></dl> <br> | ||
| Line 45: | Line 47: | ||
<dl class="imgCenter"> | <dl class="imgCenter"> | ||
| - | <dt><img alt="eforRed spectra" src="https://static.igem.org/mediawiki/2013/d/df/Braunschweig_eforRed_spektren_results.png" width=" | + | <dt><img alt="eforRed spectra" src="https://static.igem.org/mediawiki/2013/d/df/Braunschweig_eforRed_spektren_results.png" width="600" /> |
</dt> | </dt> | ||
<dd>Fig. 2: Absorption and emission spectra of eforRed.</dd></dl> <br> | <dd>Fig. 2: Absorption and emission spectra of eforRed.</dd></dl> <br> | ||
<dl class="imgCenter"> | <dl class="imgCenter"> | ||
| - | <dt><img alt="amilGFP spectra" src="https://static.igem.org/mediawiki/2013/1/11/Braunschweig_amilGFP_spektren_results.png" width=" | + | <dt><img alt="amilGFP spectra" src="https://static.igem.org/mediawiki/2013/1/11/Braunschweig_amilGFP_spektren_results.png" width="600" /> |
</dt> | </dt> | ||
<dd>Fig. 3: Absorption and emission spectra of amilGFP.</dd></dl> <br> | <dd>Fig. 3: Absorption and emission spectra of amilGFP.</dd></dl> <br> | ||
<dl class="imgCenter"> | <dl class="imgCenter"> | ||
| - | <dt><img alt="amilGFP | + | <dt><img alt="amilGFP eforRed Fluorescence" src="https://static.igem.org/mediawiki/2013/9/90/Braunschweig_Fluoreszenzaufnahme_amilGFP_eforRed.png" width="300" /> |
</dt> | </dt> | ||
| - | <dd>Fig. 4: Bacteria expressing eforRed and amilGFP can clearly be distinguished in fluorescence microscopy.</dd></dl> <br> | + | <dd>Fig. 4: Bacteria expressing eforRed and amilGFP can clearly be distinguished in fluorescence microscopy.</dd></dl> <br></p> |
| - | </ | + | |
| + | <img alt="grey line" src="https://static.igem.org/mediawiki/2013/4/4c/Braunschweig_grey_line.png" width="850" height="1" vspace="20"/> | ||
| + | |||
| + | <p style="font-size:14px; font-weight:bold; text-decoration:none; border:none; color:#be1e3c">2. The inducibility of an ampicillin resistance was shown</p> | ||
| + | |||
| + | <p>We were able to induce cell growth on ampicillin supplemented medium by addition of synthetic inducers. The expression of beta-lactamase in BBa_K1073034 and BBa_K1073035 is dependent on the induction of the promoters Plas and Prhl. The corresponding transcription activator (LasR and RhlR) is coded in each construct and expressed constitutively. Upon addition of the inducers (N-3-oxododecanoyl homoserine lactone for LasR and N-butyryl homoserine lactone for RhlR) cell growth in ampicillin containing medium was enabled whereas no significant growth was observed in control cultures (fig. 5). | ||
| + | <dl class="imgCenter"> | ||
| + | <dt><img alt="Induction of Growth" src="https://static.igem.org/mediawiki/parts/thumb/e/e4/Braunschweig2013_Top10_pSB1C3_K1073035.jpg/500px-Braunschweig2013_Top10_pSB1C3_K1073035.jpg" width="400" /> | ||
| + | </dt> | ||
| + | <dd>Fig. 5: Successful induction of cell growth on medium that was supplemented with Ampicillin.<br> | ||
| + | Cell growth of <i>E. coli</i> strains bearing BBa_K1073034 and BBa_K1073035<br> | ||
| + | in Ampicillin medium was induced by addition of the corresponding inducers.<br> | ||
| + | Due to the promoter leakiness of P<sub>las</sub> 5 µM Clavulanic acid, a ß-lactamase inhibitor,<br> | ||
| + | was added to the medium to overcome background expression of ß-lactamase by BBa_K1073034. </dd></dl> <br></p> | ||
| + | |||
| + | <img alt="grey line" src="https://static.igem.org/mediawiki/2013/4/4c/Braunschweig_grey_line.png" width="850" height="1" vspace="20"/> | ||
| + | |||
| + | <p style="font-size:14px; font-weight:bold; text-decoration:none; border:none; color:#be1e3c">3. Successful production of inducers with BBa_K1073034 and BBa_K1073035</p> | ||
| + | |||
| + | <p>Using specific homoserine lactone (HSL) reporter strains, we were able to confirm presence of inducers in culture-supernatant of <i>E. coli</i> strains bearing the devices BBa_K1073034 and BBa_K1073035. The HSL reporter strains make use of plasmids that have a luciferase cloned behind inducible promoters. When the corresponding inducers are present in the medium, the reporter strains develop a bioluminescence that can easily be detected (fig. 6). | ||
| + | |||
| + | <dl class="imgCenter"> | ||
| + | <dt><img alt="Bioluminescence" src="https://static.igem.org/mediawiki/2013/7/7d/Braunschweig_Biolumineszenz.png" width="600" /> | ||
| + | </dt> | ||
| + | <dd>Fig. 6: HSL were detected in culture supernatants using specific repoter strains.<br> | ||
| + | The reporter strains were cultivated in sterile filtered supernatant of<br> <i>E. coli</i> JM109::pSB1C3-BBa_K1073034 and <i>E. coli</i> Top10F’::pSB1C3-BBa_K1073035.<br> After 3 h cultivation bioluminescence was measured.</dd></dl> <br></p> | ||
| + | |||
| + | <img alt="grey line" src="https://static.igem.org/mediawiki/2013/4/4c/Braunschweig_grey_line.png" width="850" height="1" vspace="20"/> | ||
| + | |||
| + | <p style="font-size:14px; font-weight:bold; text-decoration:none; border:none; color:#be1e3c">4. Crossinduction of the constructs BBa_K1073034 and BBa_K1073035 was proven.</p> | ||
| + | <p> The crossinduction of beta-lactamase in E. coli strains bearing the constructs BBa_K1073034 and BBa_K1073035 was proven (fig.7). | ||
| + | <dl class="imgCenter"> | ||
| + | <dt><img alt="Agar diffusion test" src="https://static.igem.org/mediawiki/2013/7/72/Braunschweig_Agardiffusion.png" width="600" /> | ||
| + | </dt> | ||
| + | <dd>Fig. 7: Successful induction of cell growth on plates that were supplemented with ampicillin.<br> Cell growth of <i>E. coli</i> strains bearing BBa_K1073034 and BBa_K1073035<br> on ampicillin supplemented medium was induced in agar diffusion tests.<br> <i>E. coli</i> strains bearing the constructs BBa_K107303 and BBa_K1073035 were plated on<br> agar plates supplemented with ampicillin. Filter platelets soaked with sterile filtered supernatant<br> of the complementing strain were applied to the agar plates. </dd></dl> <br></p> | ||
| + | |||
</div> | </div> | ||
| + | |||
Revision as of 21:40, 4 October 2013

Results
To establish a stable microbial consortium our engineered constructs consist of different operation units. We demonstrated the functionality of each unit by individual experiments:
1. Reporter-chromoproteins were successfully integrated into the final constructs
The included reporter-chromoproteins were visible to the naked eye in less than 24 h during incubation on agar plates or in liquid culture (fig. 1). The use of chromoproteins as reporter facilitated the distinction of the different bacterial strains we intended to cultivate in co-culture. Featured chromoproteins were amilGFP, eforRed and aeBlue which developed a yellowish, bright pink and dark blue color.

- Fig. 1: Successful integration of reporter-chromoproteins into our constructs.
Liquid overnight cultures expressing the chromoproteins amilGFP (left), eforRed (middle), aeBlue (right).
However, eforRed and amilGFP can also be detected by fluorescence (fig. 2-4).

- Fig. 2: Absorption and emission spectra of eforRed.

- Fig. 3: Absorption and emission spectra of amilGFP.

- Fig. 4: Bacteria expressing eforRed and amilGFP can clearly be distinguished in fluorescence microscopy.
2. The inducibility of an ampicillin resistance was shown
We were able to induce cell growth on ampicillin supplemented medium by addition of synthetic inducers. The expression of beta-lactamase in BBa_K1073034 and BBa_K1073035 is dependent on the induction of the promoters Plas and Prhl. The corresponding transcription activator (LasR and RhlR) is coded in each construct and expressed constitutively. Upon addition of the inducers (N-3-oxododecanoyl homoserine lactone for LasR and N-butyryl homoserine lactone for RhlR) cell growth in ampicillin containing medium was enabled whereas no significant growth was observed in control cultures (fig. 5).

- Fig. 5: Successful induction of cell growth on medium that was supplemented with Ampicillin.
Cell growth of E. coli strains bearing BBa_K1073034 and BBa_K1073035
in Ampicillin medium was induced by addition of the corresponding inducers.
Due to the promoter leakiness of Plas 5 µM Clavulanic acid, a ß-lactamase inhibitor,
was added to the medium to overcome background expression of ß-lactamase by BBa_K1073034.
3. Successful production of inducers with BBa_K1073034 and BBa_K1073035
Using specific homoserine lactone (HSL) reporter strains, we were able to confirm presence of inducers in culture-supernatant of E. coli strains bearing the devices BBa_K1073034 and BBa_K1073035. The HSL reporter strains make use of plasmids that have a luciferase cloned behind inducible promoters. When the corresponding inducers are present in the medium, the reporter strains develop a bioluminescence that can easily be detected (fig. 6).
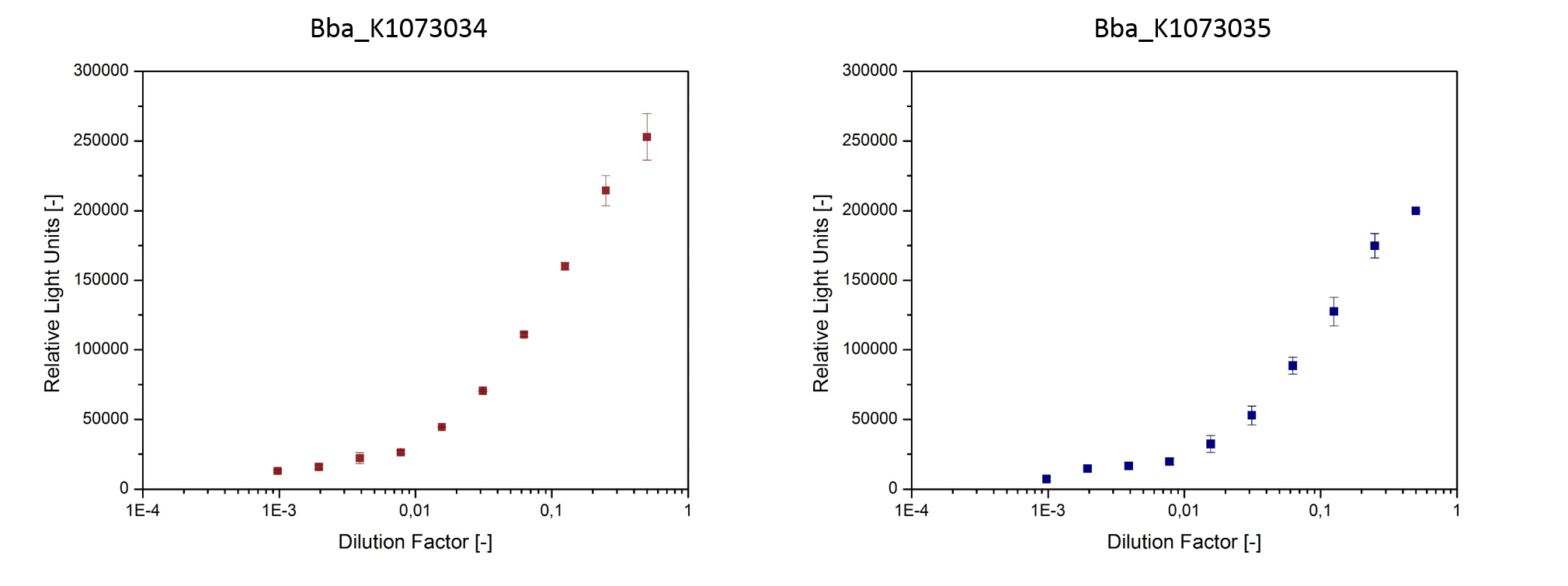
- Fig. 6: HSL were detected in culture supernatants using specific repoter strains.
The reporter strains were cultivated in sterile filtered supernatant of
E. coli JM109::pSB1C3-BBa_K1073034 and E. coli Top10F’::pSB1C3-BBa_K1073035.
After 3 h cultivation bioluminescence was measured.
4. Crossinduction of the constructs BBa_K1073034 and BBa_K1073035 was proven.
The crossinduction of beta-lactamase in E. coli strains bearing the constructs BBa_K1073034 and BBa_K1073035 was proven (fig.7).

- Fig. 7: Successful induction of cell growth on plates that were supplemented with ampicillin.
Cell growth of E. coli strains bearing BBa_K1073034 and BBa_K1073035
on ampicillin supplemented medium was induced in agar diffusion tests.
E. coli strains bearing the constructs BBa_K107303 and BBa_K1073035 were plated on
agar plates supplemented with ampicillin. Filter platelets soaked with sterile filtered supernatant
of the complementing strain were applied to the agar plates.
Our sponsors

 "
"
