Team:Heidelberg/Templates/DelH week14
From 2013.igem.org
(→Generation of DelH Plasmid 28-07) |
m |
||
| Line 99: | Line 99: | ||
<br/> | <br/> | ||
[[File:Heidelberg_20130802 colonyPCR.png|200px|thumb|right|'''Fig.14.1''' Gel of picked colonies with insert DelH-BB (loaded 20 µL of PCR) <br> ''l1:'' 1, ''l2:'' 2,''l3:'' 3, ''l4:'' 2log,''l5:'' 4, ''l6:'' 5,''l7:'' 6 <br> no bands = no colony positive]] | [[File:Heidelberg_20130802 colonyPCR.png|200px|thumb|right|'''Fig.14.1''' Gel of picked colonies with insert DelH-BB (loaded 20 µL of PCR) <br> ''l1:'' 1, ''l2:'' 2,''l3:'' 3, ''l4:'' 2log,''l5:'' 4, ''l6:'' 5,''l7:'' 6 <br> no bands = no colony positive]] | ||
| - | + | ||
Gel does not show any band. | Gel does not show any band. | ||
:=> None of the analyzed colonies is positive. | :=> None of the analyzed colonies is positive. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
====Colony-PCR Conditions CP.W14.B==== | ====Colony-PCR Conditions CP.W14.B==== | ||
Picked 20 colonies per plate and 5 colonies per PCR tube. | Picked 20 colonies per plate and 5 colonies per PCR tube. | ||
| Line 148: | Line 149: | ||
Expected band: 663 bp | Expected band: 663 bp | ||
<br/> | <br/> | ||
| - | [[File:Heidelberg_20130803 | + | [[File:Heidelberg_20130803 colonyPCR4-6.png|200px|thumb|right|'''Fig.14.3''' Gel of picked colonies with insert DelH-BB (loaded 20 µL of PCR) <br>''l1:'' 2log, ''l2-5:'' 20 picked colonies from plate 4: every PCR tube has 5 colonies, ''l6-9:''20 picked colonies from plate 5every PCR tube has 5 colonies,''l10-14:''20 picked colonies from plate 6 every PCR tube has 5 colonies<br> bands on lanes 4a, 4b, 6a, 6b, 6d at the expected 663 bp, so the next step is making PCRs of 1 colony per PCR of these probes]] |
| - | + | <div class="tright" style="clear:none">[[File:Heidelberg_20130803 colonyPCR1-3.png|200px|thumb|right|'''Fig.14.2''' Gel of picked colonies with insert DelH-BB (loaded 20 µL of PCR) <br> ''l1-3:''3 picked colonies from plate 1, every PCR tube has 5 colonies, ''l4-7:''4 picked colonies from plate 2, every PCR tube has 5 colonies,''l8:'' 2log,''l9-13:''4 picked colonies from plate 3 every PCR tube has 5 colonies <br> bands on lanes 3b, 3c, 3d, 4a, 4b, 6a, 6b, 6d at the expected 663 bp, so the next step is making PCRs of 1 colony per PCR of these probes]]</div> | |
| - | + | ||
Gel shows bands for b, 3c, 3d, 4a, 4b, 6a, 6b, 6d at the expected 663 bp. | Gel shows bands for b, 3c, 3d, 4a, 4b, 6a, 6b, 6d at the expected 663 bp. | ||
:=> Make single colony-PCRs. | :=> Make single colony-PCRs. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
====Colony-PCR Conditions CP.W14.C==== | ====Colony-PCR Conditions CP.W14.C==== | ||
1 µl of colony ON culture picked | 1 µl of colony ON culture picked | ||
| Line 198: | Line 200: | ||
Expected band: 663 bp | Expected band: 663 bp | ||
<br/> | <br/> | ||
| - | [[File:Heidelberg_20130803 | + | [[File:Heidelberg_20130803 colonyPCR6b.png|200px|thumb|right|'''Fig.14.5''' Gel of picked colonies with insert DelH-BB (loaded 20 µL of PCR) <br>'l1:'' 2log, ''l2-6:'' 5 seperate colonies which were together in 6b PCR earlier,<br> no bands = no colony positive]] |
| - | + | <div class="tright" style="clear:none">[[File:Heidelberg_20130803 colonyPCR3c-4a.png|200px|thumb|right|'''Fig.14.4''' Gel of picked colonies with insert DelH-BB (loaded 20 µL of PCR) <br>'l1:'' 2log, ''l2-6:'' 5 seperate colonies which were together in 3a PCR earlier, ''l7-11:'' 5 seperate colonies which were together in 4b PCR earlier, <br> no bands = no colony positive]]</div> | |
| - | + | ||
| + | |||
:=> No band is visible. There should be a band at at least one of each sample (for example 6B), because in the previous PCR we observed a band. | :=> No band is visible. There should be a band at at least one of each sample (for example 6B), because in the previous PCR we observed a band. | ||
<br/> | <br/> | ||
Because of the explained reason above we incubated 40 colonies seperately which might be positive ON at 37°C in 200 µl LB Amp. 200 µl of the ON cultire was mixed with 1.8 ml LB Amp for five colonies of each probe: 3b, 3c, 3d, 4a, 4b, 6a, 6b, 6d. | Because of the explained reason above we incubated 40 colonies seperately which might be positive ON at 37°C in 200 µl LB Amp. 200 µl of the ON cultire was mixed with 1.8 ml LB Amp for five colonies of each probe: 3b, 3c, 3d, 4a, 4b, 6a, 6b, 6d. | ||
<br/> | <br/> | ||
| + | <div style="clear:both"></div> | ||
Revision as of 11:03, 25 October 2013
Contents |
29-07 - 04-08-13
Generation of DelH Plasmid 28-07
Purification of Gibson Mix
10 µl of the Gibson mix were isopropanol purified as described in the protocol.
5 µl of the Gibson reaction were mixed with 10 µl ddH2O.
Electroporation
- Electroporation into freshly prepared electrocompetent cell following the protocol.
| E1 | E2 | E3 |
|---|---|---|
| 5 µl of Gibson + 10 µl H2O | 10 µl of isoprop purified Gibson assembly | no DNA |
- Grow 0.5 h with 400 µl SOC at 37°C
- Add 9 ml LB Amp and incubate 3 h at 37°C
- Incubate ON at 37°C on LB Amp plate as well as LB (no Amp) for control purposes
Result
There were few colonies on the plates. Red colonies were picked and screenied by colony-PCR, but none of them was positive. The Gibson assembly has to be repeated.
Generation of DelH Plasmid 01-08
Gibson Assembly
| Fragment | Concentraion | Amount used [µl] (2 vials) | Amount used [µl] |
|---|---|---|---|
| G0 | 0,68 ng/µl | 9 | 6 |
| BB (8.8) | 22 ng/µl | 1 | 4 |
| Gibson Master Mix | 2x | 10 | 10 |
- Incubated 1 h at 50 °C in thermocycler
Electroporation
| Electroporation # | Mix # | Amount of Mix [µl] | ddH2O [µl] | Isopropanol | Amount electroporated [µl] |
|---|---|---|---|---|---|
| 1 | 1 | 10 | 20 | - | 1 |
| 2 | 1 | 10 | 20 | - | 14 |
| 3 | 1 | 30 | - | yes | 20 |
| 4 | 2 | 5 | 10 | - | 1 |
| 5 | 2 | 5 | 10 | - | 29 |
| 6 | 2 | 15 | - | yes | 20 |
| 7 | - | - | 10 | - | 10 |
- Plated on one LB agar plate each
- Stored ON at 37°C
Colony-PCR Conditions CP.W14.A
| Reagent | Electroporation 1 | Electroporation 2 | Electroporation 3 | Electroporation 4 | Electroporation 5 | Electroporation 6 |
|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 1 | 2 | 3 | 4 | 5 | 6 |
| Template | 5 colonies of E1 | 5 colonies of E2 | 5 colonies of E3 | 5 colonies of E4 | 5 colonies of E5 | 5 colonies of E6 |
| Primer fw 10 µM | 1 µl VF2 | 1 µl VF2 | 1 µl VF2 | 1 µl VF2 | 1 µl VF2 | 1 µl VF2 |
| Primer rev 10 µM | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev | 1 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 8 µl | 8 µl | 8 µl | 8 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Gel does not show any band.
- => None of the analyzed colonies is positive.
Colony-PCR Conditions CP.W14.B
Picked 20 colonies per plate and 5 colonies per PCR tube.
| Reagent | Electr. 1 | Electr. 1 | Electr. 1 | Electr. 2 | Electr. 2 | Electr. 2 | Electr. 2 | Electr. 3 | Electr. 3 | Electr. 3 | Electr. 3 | Electr. 4 | Electr. 4 | Electr. 4 | Electr. 4 | Electr. 5 | Electr. 5 | Electr. 5 | Electr. 5 | Electr. 6 | Electr. 6 | Electr. 6 | Electr. 6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 1a | 1b | 1c | 2a | 2b | 2c | 2d | 3a | 3b | 3c | 3d | 4a | 4b | 4c | 4d | 5a | 5b | 5c | 5d | 6a | 6b | 6c | 6d |
| Template | 5 colonies of E1 | 5 colonies of E1 | 5 colonies of E1 | 5 colonies of E2 | 5 colonies of E2 | 5 colonies of E2 | 5 colonies of E2 | 5 colonies of E3 | 5 colonies of E3 | 5 colonies of E3 | 5 colonies of E3 | 5 colonies of E4 | 5 colonies of E4 | 5 colonies of E4 | 5 colonies of E4 | 5 colonies of E5 | 5 colonies of E5 | 5 colonies of E5 | 5 colonies of E5 | 5 colonies of E6 | 5 colonies of E6 | 5 colonies of E6 | 5 colonies of E6 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp

l1: 2log, l2-5: 20 picked colonies from plate 4: every PCR tube has 5 colonies, l6-9:20 picked colonies from plate 5every PCR tube has 5 colonies,l10-14:20 picked colonies from plate 6 every PCR tube has 5 colonies
bands on lanes 4a, 4b, 6a, 6b, 6d at the expected 663 bp, so the next step is making PCRs of 1 colony per PCR of these probes

l1-3:3 picked colonies from plate 1, every PCR tube has 5 colonies, l4-7:4 picked colonies from plate 2, every PCR tube has 5 colonies,l8: 2log,l9-13:4 picked colonies from plate 3 every PCR tube has 5 colonies
bands on lanes 3b, 3c, 3d, 4a, 4b, 6a, 6b, 6d at the expected 663 bp, so the next step is making PCRs of 1 colony per PCR of these probes
Gel shows bands for b, 3c, 3d, 4a, 4b, 6a, 6b, 6d at the expected 663 bp.
- => Make single colony-PCRs.
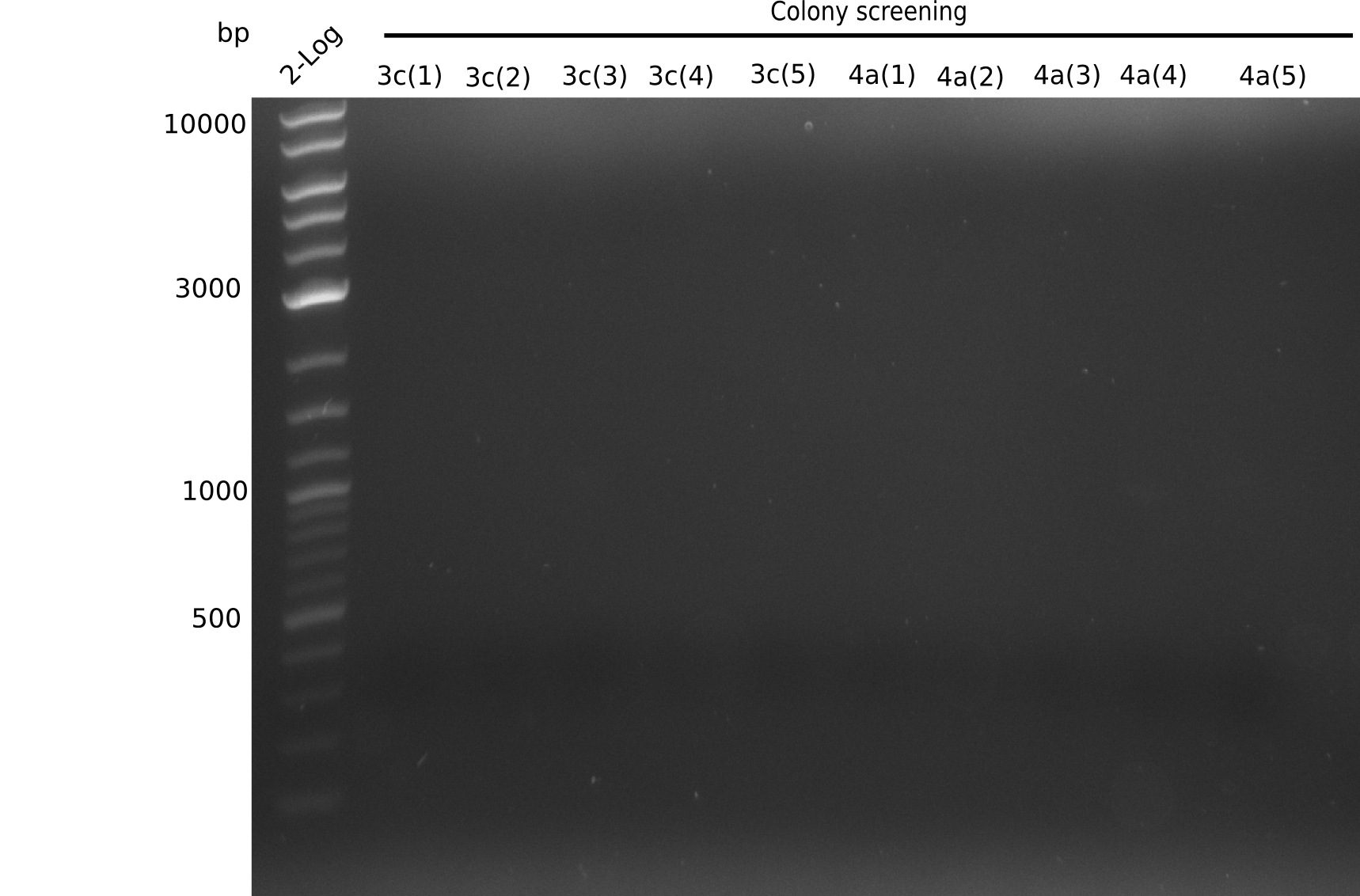
Colony-PCR Conditions CP.W14.C
1 µl of colony ON culture picked
| Reagent | 3c | 3c | 3c | 3c | 3c | 4a | 4a | 4a | 4a | 4a | 6b | 6b | 6b | 6b | 6b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 3c 1 | 3c 2 | 3c 3 | 3c 4 | 3c 5 | 4a 1 | 4a 2 | 4a 3 | 4a 4 | 4a 5 | 6b 1 | 6b 2 | 6b 3 | 6b 4 | 6b 5 |
| Template (1 µl) | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E3c | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E4a | 1 colonies of E6b | 1 colonies of E6b | 1 colonies of E6b | 1 colonies of E6b | 1 colonies of E6b |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev | 2 µl Screen_delH_rev |
| Dream-Taq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 5 µl | 5 µl | 5µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5 µl | 5µl | 5 µl | 5 µl |
| Cycles | Temperature DelH-G0 [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
- => No band is visible. There should be a band at at least one of each sample (for example 6B), because in the previous PCR we observed a band.
Because of the explained reason above we incubated 40 colonies seperately which might be positive ON at 37°C in 200 µl LB Amp. 200 µl of the ON cultire was mixed with 1.8 ml LB Amp for five colonies of each probe: 3b, 3c, 3d, 4a, 4b, 6a, 6b, 6d.
 "
"