Team:Penn/AssayOverview
From 2013.igem.org
(Difference between revisions)
| Line 88: | Line 88: | ||
<b>MaGellin Workflow.</b> The workflow for screening new fusion proteins with the one plasmid MaGellin bacterial system is as follows:<br> | <b>MaGellin Workflow.</b> The workflow for screening new fusion proteins with the one plasmid MaGellin bacterial system is as follows:<br> | ||
| - | < | + | <embed src = "https://swiffypreviews.googleusercontent.com/view/o/9e04d263-21c1-4e49-bff0-af44c660434c/2013_iGEM_FinalAnimation.html""></embed> |
<>Assemble</b> the MaGellin backbone together with a DNA-binding protein and target sequence of your choosing. | <>Assemble</b> the MaGellin backbone together with a DNA-binding protein and target sequence of your choosing. | ||
Revision as of 06:53, 28 October 2013
Assay Overview
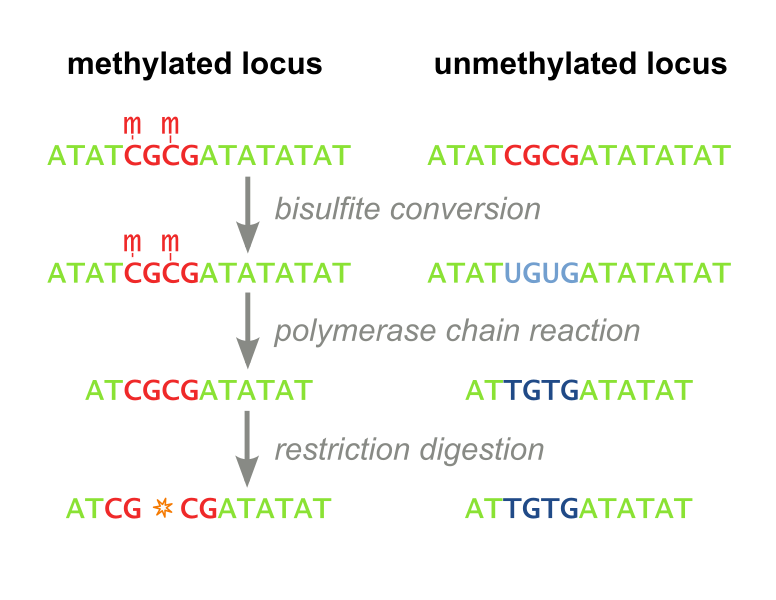

The MaGellin Methylation Assay

- CpG Methyltransferase (M.SssI) with a generic linker sequence in the cloning site. For a working fusion protein and assay, only a DNA binding domain must be cloned into the plasmid. This inherently standardizes MaGellin and lessens the time a user of the assay must spend cloning.
- Multiple cloning site downstream of T7 promoter for orthogonal expression of fusion protein in T7 Express competent E. coli.
- Cloning site for a smaller DNA sequence, specific to the fusion protein being screened – named the “target site”, where the protein will bind. This can be the binding site for a CRISPR-Cas, TALE, Zinc Finger, or transcription factor
- AvaI restriction site 4 bases downstream of the target site – the AvaI restriction enzyme is blocked by methylated CpG sites, thus screening for site specific methylation becomes equivalent to screening for AvaI digestion
- AvaI restriction site sufficiently further downstream of the target site – named the off-target site. This site screens for non-specific DNA methylation as it is spatially removed from where the fusion protein binds to the plasmid.
- XbaI site for linearization of the plasmid. Linearizing the plasmid simplifies analysis of the AvaI digestion by gel electrophoresis.
- Validated bisulfite conversion primer binding sites, so users do not need to go through the time-consuming primer design process if they choose to fortify MaGellin’s results with bisulfite sequencing results
- sgRNA cloning site for users who want to target a CRISPR-Cas binding domain. The sgRNA is constitutively expressed and can be swapped by restriction digest.
- Validated bisulfite conversion primers for users who choose to advance to bisulfite sequencing, for even higher resolution in detecting methylation, after proving their enzyme’s efficacy with our MaGellin assay.
- Kanamycin resistance as a selection marker
- T7 RNA Polymerase in the lac operon allows us to turn on expression of fusion protein after induction with IPTG
- In the T7 Express cell line, genes for several restriction enzymes known to target methylated DNA are knocked out (McrA-, McrBC-, EcoBr-m-, Mrr-). This ensures that our assay plasmid is not cleaved in vivo. Results are difficult, if not impossible, to interpret in the commonly used BL21 cell line.
<>Assemble the MaGellin backbone together with a DNA-binding protein and target sequence of your choosing.
- Forward: CAGGAGGAATTC[ATG] (add start codon only if not included in gene).
- Reverse: CTCTAGAAGCGGC (make sure to remove the stop codon).
- Transform the completed MaGellin plasmid into T7 Express.
- Induce culture with 1 mM IPTG.
- Incubate in a shaker at 37C for 5 hours.
- Miniprep to isolate the plasmid.
- Digest 600 ng of miniprep DNA in a 15 uL reaction with 10 U of both XbaI and AvaI.
- Incubate reaction for 1 hour at 37C.
- Look for 3 distinct band patterns that correspond to specific and interpretable methylation outcomes.
- The presence of large one band corresponds to non-site-specific DNA methylation (AvaI was blocked at both the target and off target sites, and thus only XbaI cut the plasmid)
- The presence of two bands corresponds to site-specific DNA methylation (AvaI was only blocked at the target site, thus AvaI cut in the off target site and XbaI cut the plasmid)
- The presence of three bands corresponds to no DNA methylation – or an inactive fusion protein (AvaI was not blocked at either the target or off target sites and XbaI cut the plasmid)

 "
"