Team:DTU-Denmark/Notebook/30 July 2013
From 2013.igem.org
(Difference between revisions)
(→PCR) |
|||
| Line 20: | Line 20: | ||
*pZE21::GFP SF and pZE21::RFP in duplicates with primers pair 13 on 55C and extension time 3:00. | *pZE21::GFP SF and pZE21::RFP in duplicates with primers pair 13 on 55C and extension time 3:00. | ||
*cycAX in triplicates with primer pair 33 2 samples on 62C and 3:30 extension and 1 sample on 55C and extension 3:00. | *cycAX in triplicates with primer pair 33 2 samples on 62C and 3:30 extension and 1 sample on 55C and extension 3:00. | ||
| - | *None negative was made for each containing same things as real samples just MQ instead of template DNA. | + | *None negative was made for each containing same things as real samples just MQ instead of template DNA. |
| + | ===PCR of Nir on genomic DNA=== | ||
| + | |||
| + | Two kinds of templates were used for this PCR: | ||
| + | * isolated genomic DNA from ''P. aeruginosa'' | ||
| + | * isolated genomic DNA from ''P. aeruginosa'' digested with HindIII | ||
| + | |||
| + | names of samples: | ||
| + | * 1,2 - primers 41a, 41b - undigested genomic DNA - part 1 of Nir | ||
| + | * 3,4 - primers 42a, 42b - undigested genomic DNA - part 2 of Nir | ||
| + | * 5,6 - primers 41a, 41b - digested genomic DNA - part 1 of Nir | ||
| + | * 7,8 - primers 42a, 42b - digested genomic DNA - part 2 of Nir | ||
| + | * 9 - negative, primers 41a, 41b | ||
| + | * 10 - negative, primers 42a, 42b | ||
| + | |||
| + | ===Miniprep=== | ||
| + | |||
| + | Isolated plasmids from yesterdays ON cultures of AMO colony 2 and colony 3 using ethanol precipitation (see modified protocol). | ||
==Results== | ==Results== | ||
Revision as of 12:45, 30 July 2013
30 July 2013
Contents |
lab 208
Main purpose
Make PCR on Nir with genomic DNA template. Amplify the pZE21 backbone with USER-primers for construction of the arabinose inducible system. Put His-tag on the cycAX construct.
Who was in the lab
Gosia, Kristian, Henrike, Julia
Procedure
PCR
- pZE21::GFP SF and pZE21::RFP in duplicates with primers pair 13 on 55C and extension time 3:00.
- cycAX in triplicates with primer pair 33 2 samples on 62C and 3:30 extension and 1 sample on 55C and extension 3:00.
- None negative was made for each containing same things as real samples just MQ instead of template DNA.
PCR of Nir on genomic DNA
Two kinds of templates were used for this PCR:
- isolated genomic DNA from P. aeruginosa
- isolated genomic DNA from P. aeruginosa digested with HindIII
names of samples:
- 1,2 - primers 41a, 41b - undigested genomic DNA - part 1 of Nir
- 3,4 - primers 42a, 42b - undigested genomic DNA - part 2 of Nir
- 5,6 - primers 41a, 41b - digested genomic DNA - part 1 of Nir
- 7,8 - primers 42a, 42b - digested genomic DNA - part 2 of Nir
- 9 - negative, primers 41a, 41b
- 10 - negative, primers 42a, 42b
Miniprep
Isolated plasmids from yesterdays ON cultures of AMO colony 2 and colony 3 using ethanol precipitation (see modified protocol).
Results
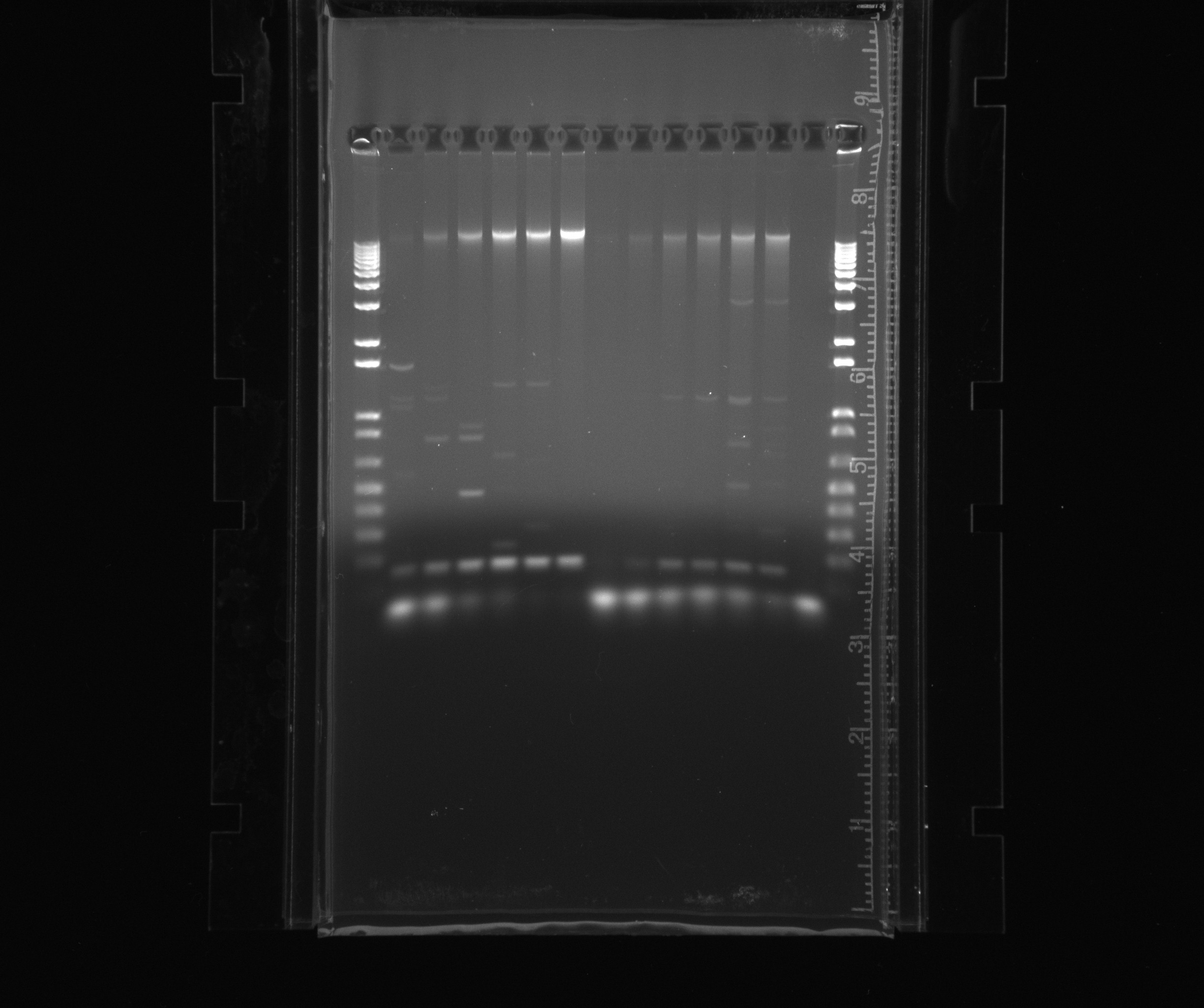
Gel on PCR from yesterday
- 1kb ladder
- Nir 1uL genomic template
- Nir 3uL genomic template
- Nir 7uL....
- Nir 10uL....
- Nir 12uL....
- Nir 15uL....
- Nir 1uL HindIII treated genomic template
- Nir 3uL HindIII treated genomic template
- Nir 7uL....
- Nir 10uL....
- Nir 12uL....
- Nir 15uL....
- Negative control
- 1kb ladder
Conclusion
No right bands where acquired from PCR-amplification with genomic DNA template.
Navigate to the Previous or the Next Entry
 "
"