Tim Baker
From 2013.igem.org
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <div id="header">{{Template:Team:Yale2013/Templates/Header}}</div> | ||
| + | |||
'''[https://2013.igem.org/Team:Yale/Notebook Return to Notebooks]''' | '''[https://2013.igem.org/Team:Yale/Notebook Return to Notebooks]''' | ||
Revision as of 20:08, 2 August 2013
Contents |
July 26, 2013
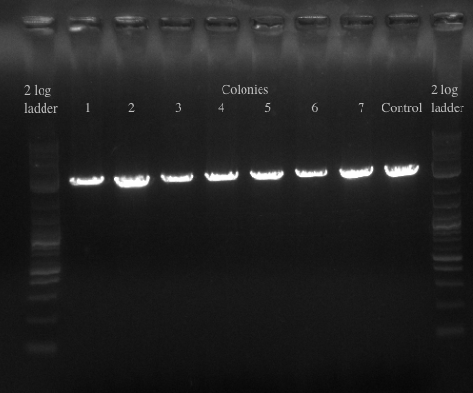
- Re-made the template from the 8 cultures of cells and then PCR amplified with primers 49 and 125 which have homology to 21B
- Ran the PCR for all of the colonies to confirm the presence of the gene
July 25, 2013
- Checked the double stranded Recombination plates from yesterday
- Picked the 7 colonies from the EcNR1 positive plate into 3 mL of LB min medium
- Let grow for a few hours, then used them as a template to screen at 21B
- Will either get a ~5.5 kb fragment (PHA/Pct + kanR) if it worked or a ~2 kb fragment (tolC) if it didn't
- Results:
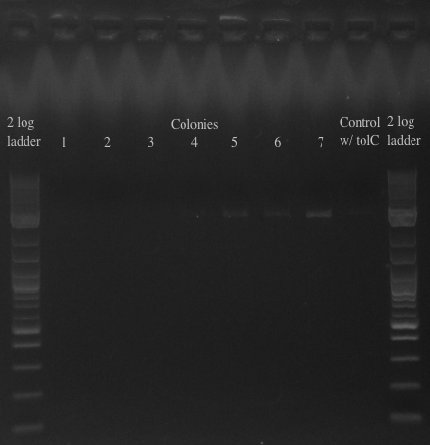
- All of the bands were not very clear, probably a result of various factors, including using a different polymerase, mixing in the primers with the mastermix, not letting the cells grow up for long enough, and the gel comb set at an unusual angle. Will redo tomorrow
July 24, 2013
- Plated the cells on Kanamycin plates
- 3 different plates for each strain that was used in the recombination
- Negative control with water as the DNA
- 50 uL of the 1 mL culture after ~15 hours of growth with 100 ng of DNA in 50 uL of water
- Spun down the cells after ~15 hours of growth from the rest of the culture and resuspended in 50 uL of water with 100 ng of DNA in 50 uL of water
- Plan on screening the 21B sight with primers tomorrow
July 23, 2013
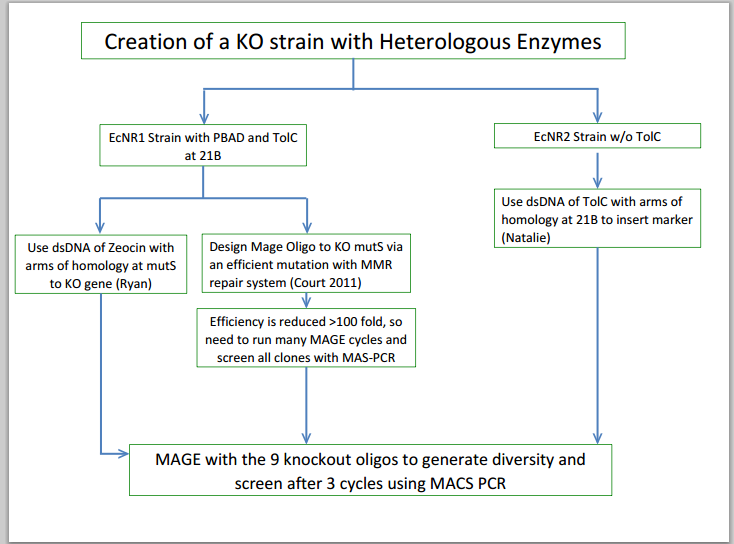
- Grew up strains of EcNR1 mutS zeo tolC 21B and A3 4/4 which Andrew finished getting all of the knockouts on
- Strains took an unusually long time to reach midlog
- Induced cells at 42 degrees celsius and continued with the double stranded recombination protocol that Natalie provided
- Let the cells recover overnight because they have not been growing very efficiently
July 22, 2013
- Updated the Weekly Meeting presentation for the week of 7/22
- Meeting went well, Dellaporta suggested running a gradient PCR for the PHA/Pct + kanR fragment
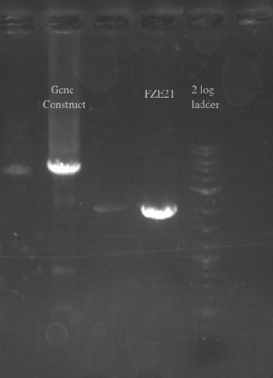
- Checked the EcNR1... cells that were transformed with PHA/Pct on PZE21
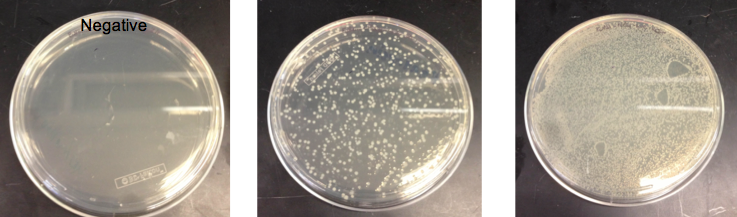
- Picked a single, medium sized, colony and started a culture for frozen stocks
- Ran a gradient PCR on the PHA/Pct + kanR fragment as suggested by Dellaporta: Temperature range was 72-65
File:Screen Shot 2013-07-24 at 12.32.31 PM.png- The Gel shows clear bands all the way across at 5.5 kb, not sure what the other bands could be considering I used only primers 49 and 125
- Gel purified all of the bands at 5.5 kb and nano-dropped to get a concentration of 66.3 ng/uL
July 19, 2013
- Creating and transforming a plasmid with PHA/Pct
- Positive Selection with kanR
- PCR amplified PHA/Pct and kanR with homology to one another
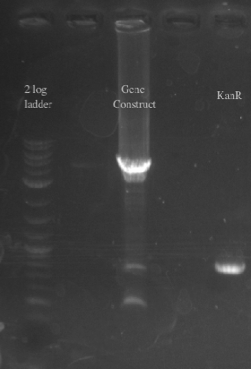
- Gibson assembled the two fragments together
- PCR amplified the Gibson assembly to increase the concentration for double stranded recombination
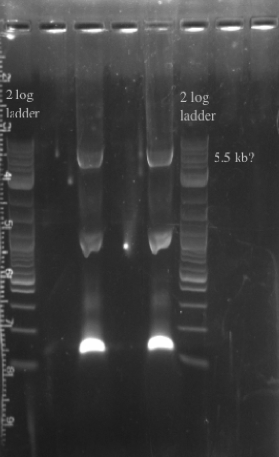
- Not sure whether the band is at 5.5 kb or not, will probably have to redo the PCR for better clarity
- Gel purified and nano-dropped to get a concentration of ~20 ng/uL
- PCR amplified PHA/Pct and kanR with homology to one another
July 18, 2013
- Ordered and received primers to amplify BB29 and BB30 without homology and then the PZE21 backbone with more homology
- Used BB29 and BB30 cells from frozen stocks as template DNA
- Primers 138-145
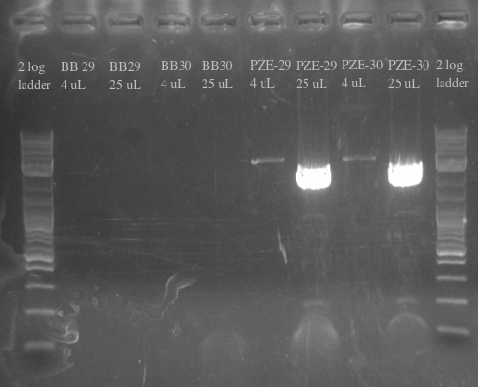
- Repeated failure to amplify the biobricks fragments suggests that there is an error in either the sequence or the shipment itself
July 17, 2013
- Ran another PCR on the Biobricks fragments and backbone to see if I could resolve the fragments using a different DNA template
- Used cells grown up overnight as the template for BB29 and BB30
- Results:
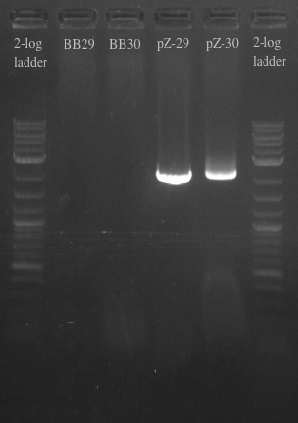
- Gel failed again pointing to something wrong with the primers
- Checked primers and re-ordered a set of primers to just amplify the BB fragments as well as amplify the backbone with longer homology for the gibson assembly
- Gel failed again pointing to something wrong with the primers
July 16, 2013
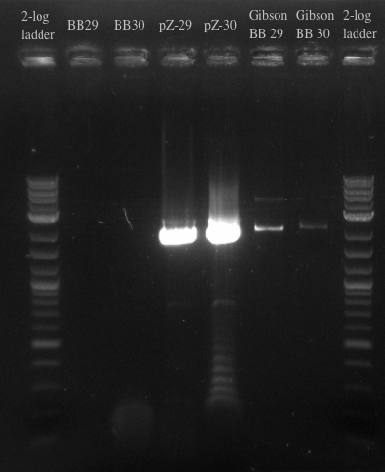
- Ran a gibson assembly and re-ran the PCR for the Biobricks fragments and the backbone on the same gel
- Results indicate that the Gibson for BB29 may have worked but there is likely something wrong with the primers for the BioBricks because the band at 900 did not show up
July 15, 2013
- Weekly meeting presentation with Farren went well: advised to forget the positive control after unpromising results
- Diluted and created working stocks of the primers for the addition of the Biobrick fragments into the expression plasmid
- Trying to create two plasmids
- Expression plasmid with PLtet-O promoter and gfp + hlya tag + terminator
- Expression plasmid with PLtet-O promoter and phasin + hlya tag + terminator
- Ran PCR gel purification to amplify the Biobricks fragments and the expression vector backbone with homology for Gibson assembly
- Nanodropped
July 10, 2013
- Continued to design sequence primers for the RBS modifications and for Knockouts
- Emailed Ken Nelson and set up a time to learn how to use the FACS Verse
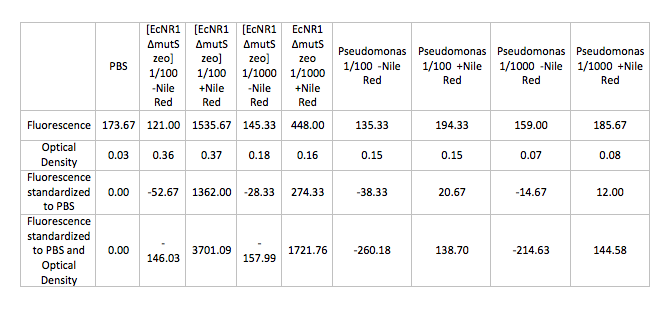
- Ran Positive control on the Pseudomonas and [EcNR1 ^mutS zeo] strains grown in Nile Red according to the following procedure:
- Back-dilute 2 overnight cultures of Pseudomonas and [EcNR1 ^mutS zeo] at concentrations of 1/100 and 1/1000 in 3mL of BE and LB min medium respectively
- Add Nile Red to the positive control samples at a concentration of 0.5 ug/mL
- Allow samples to grow in shaking incubator at 34 degrees celsius for 3-4 hours
- Spin down 1mL of cells in the microcentrifuge and wash and resuspend the pellets in PBS
- Test cells on fluorescence plate reader standardized to a PBS solution
July 9, 2013
- Received strain of Pseudomonas, created 3 cultures with Beef Extract Medium and put them in shaking incubator
- 1 culture with the entirety of the concentrated pellet in 6mL of BE Medium, 2 cultures with 100 uL of the concentrated culture in 5 mL of medium
- Plated 2 cultures (concentrate and dilute) and put them in the 32 degree incubator
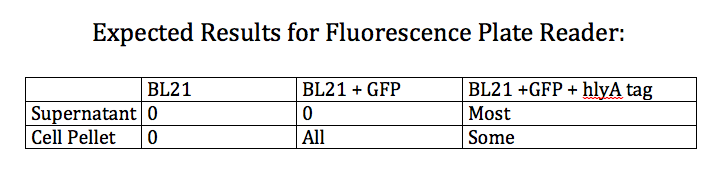
- Designed an experiment as a positive control for the effectiveness of the type 1 export system
- Designed Sequence Primers for RBS Modifications and for Knockouts
July 8, 2013
- Created 500mL of the 3 Nutrient Broth (Beef Extract Medium)
- 1.5g Meat Extract + 2.5g Peptone
- Created 500mL of the 3 Nutrient Agar (Beef Extract Agar) and poured 24 plates
- 1.5g Meat Extract + 2.5g Peptone + 7.5g Bacteriological Agar
- Designed experiment to test whether nile red can effectively stain Pseudomonas cells producing PHAs
- Test 4 groups: E. Coli cells washed with PBS grown with and without nile red and Pseudomonas cells washed with PBS grown with and without nile red (all standardized to PBS)
June 26, 2013
June 18, 2013
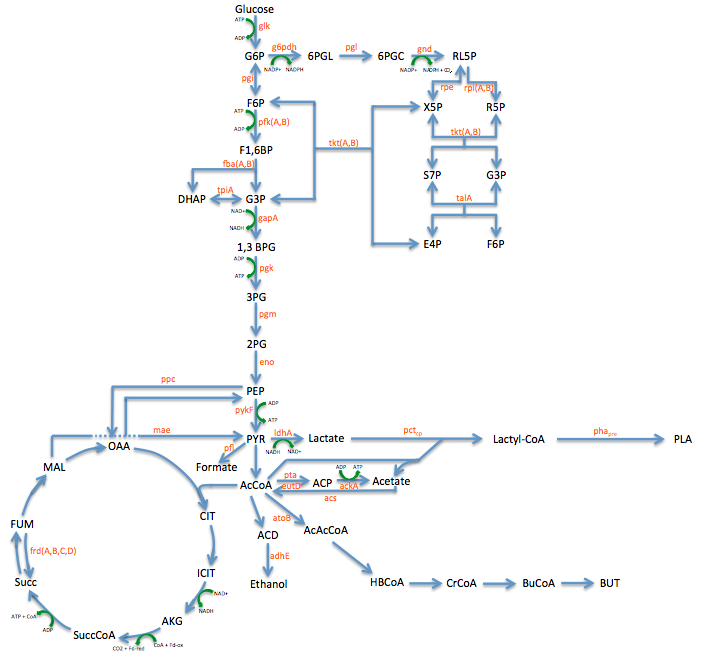
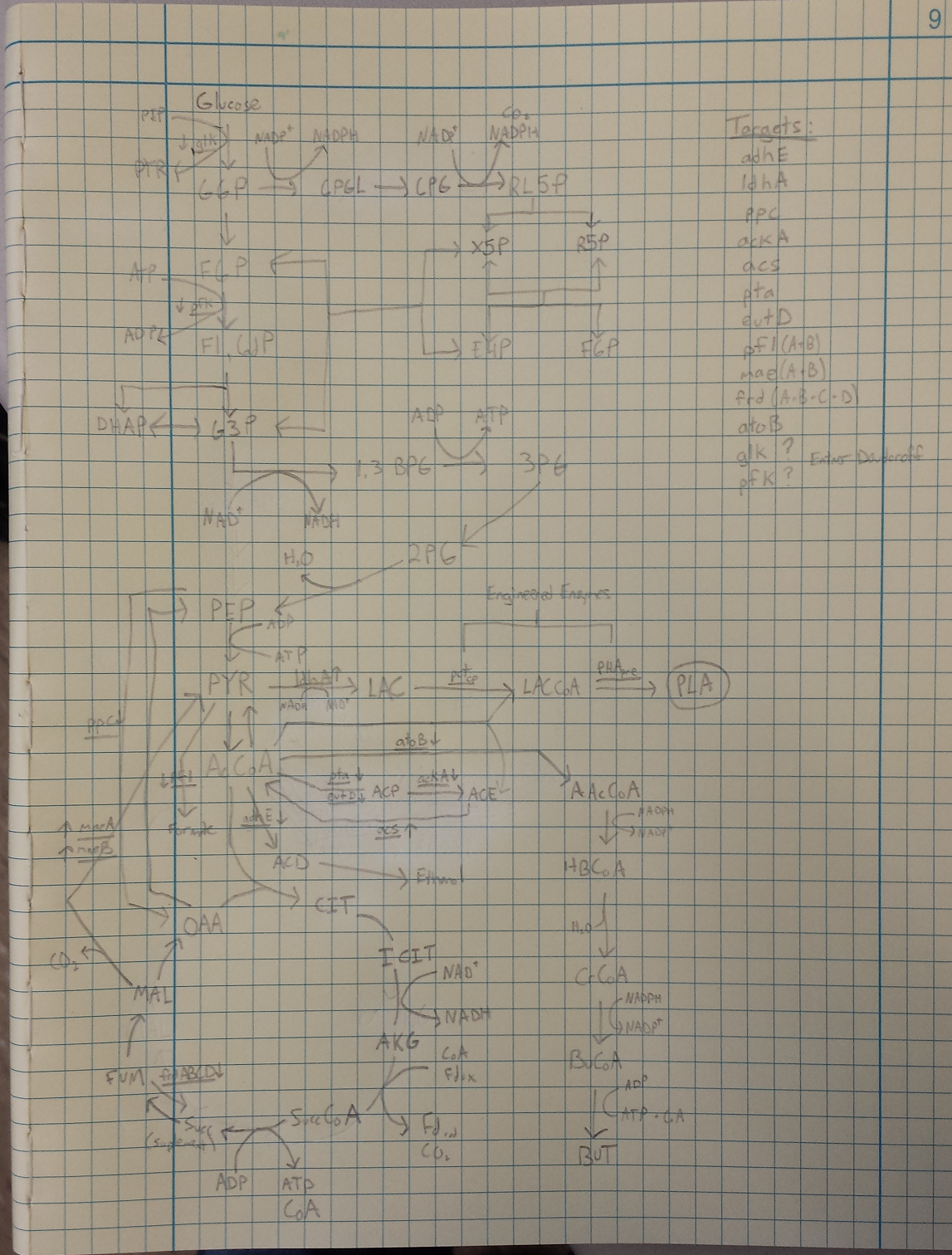
- Metabolic Engineering Graphic
June 14, 2013
- Researched metabolic engineering to increase PLA
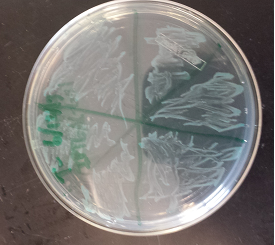
- Took pictures of the second cycle of MAGE practice
June 12, 2013
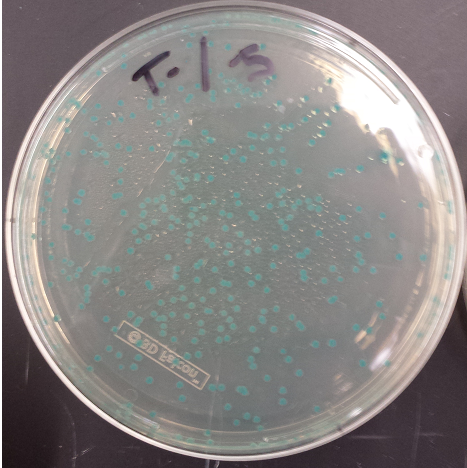
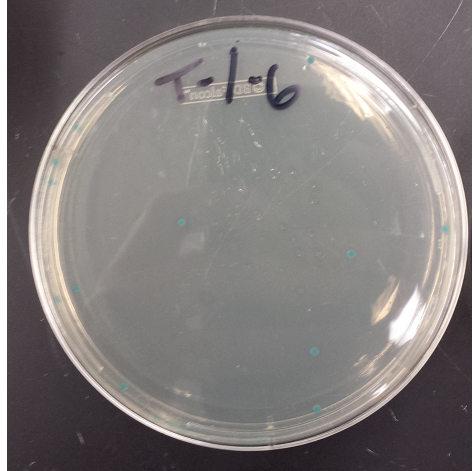
- Positive MAGE Plate
- Replated negative control MAGE colonies and they worked (initial plates went bad, due to light exposure)
 "
"