Team:DTU-Denmark/Notebook/30 August 2013
From 2013.igem.org
(Difference between revisions)
(→Procedure) |
(→PCR to amplify HAO and cycAX USER fragments) |
||
| Line 13: | Line 13: | ||
===PCR to amplify HAO and cycAX USER fragments=== | ===PCR to amplify HAO and cycAX USER fragments=== | ||
| + | |||
| + | Made 6 reactions for each, using either the extraction PCR fragment or the glycerol stock as template | ||
| + | |||
| + | {| class="wikitable" style="text-align: right" | ||
| + | ! label !! cyc gly D !! cyc gly % !! cyc gly 1uL D !! cyc ext D !! cyc ext % !! cyc ext 1uL D !! | ||
| + | |- | ||
| + | | template || glycerol stock || glycerol stock || glycerol stock || extraction PCR fragment || extraction PCR fragment || extraction PCR fragment | ||
| + | |- | ||
| + | | DMSO || 1,5 || - || 1,5 || 1,5 || - || 1,5 | ||
| + | |- | ||
| + | | MgCl2 || - || - || 1 || - || - || 1 | ||
| + | |- | ||
| + | | MQ || 1 || 2,5 || - || 1 || 2,5 || - | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | Program: | ||
| + | {| class="wikitable" style="text-align: right" | ||
| + | ! temperature !! time !! cycles | ||
| + | |- | ||
| + | | 98C || 2:00 || - | ||
| + | |- | ||
| + | | 98C || 0:20 || 36 | ||
| + | |- | ||
| + | | annealing temperature || 1:00 || 36 | ||
| + | |- | ||
| + | | 72C || extension time || 36 | ||
| + | |- | ||
| + | | 72C || 5:00 || - | ||
| + | |- | ||
| + | | 10C || hold || - | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | HAO - 59C, 3:00 | ||
| + | cycAX - 55C, 1:30 | ||
==Results== | ==Results== | ||
Revision as of 17:37, 30 August 2013
30 August 2013
Contents |
lab 208
Main purpose
Who was in the lab
Procedure
PCR to amplify HAO and cycAX USER fragments
Made 6 reactions for each, using either the extraction PCR fragment or the glycerol stock as template
| label | cyc gly D | cyc gly % | cyc gly 1uL D | cyc ext D | cyc ext % | cyc ext 1uL D | |
|---|---|---|---|---|---|---|---|
| template | glycerol stock | glycerol stock | glycerol stock | extraction PCR fragment | extraction PCR fragment | extraction PCR fragment | |
| DMSO | 1,5 | - | 1,5 | 1,5 | - | 1,5 | |
| MgCl2 | - | - | 1 | - | - | 1 | |
| MQ | 1 | 2,5 | - | 1 | 2,5 | - |
Program:
| temperature | time | cycles |
|---|---|---|
| 98C | 2:00 | - |
| 98C | 0:20 | 36 |
| annealing temperature | 1:00 | 36 |
| 72C | extension time | 36 |
| 72C | 5:00 | - |
| 10C | hold | - |
HAO - 59C, 3:00 cycAX - 55C, 1:30
Results
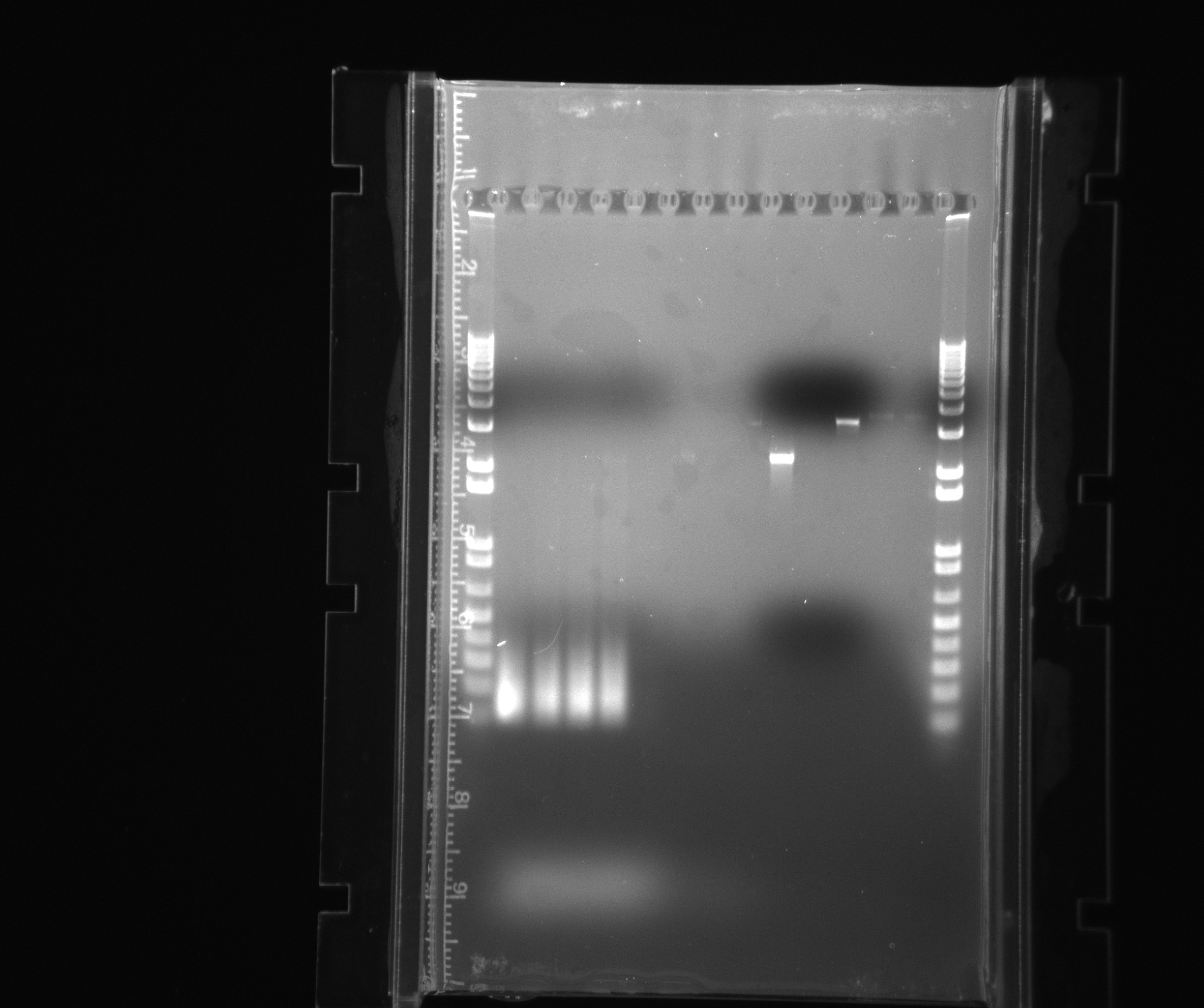
Gel
- 1 kb ladder
- psB1C3 for Nir, 2% DMSO
- psB1C3 for Nir, 2% DMSO, 1 uL 50mM MgCl2
- psB1C3 for Nir, 4% DMSO
- psB1C3 for Nir, 4% DMSO, 1 uL 50mM MgCl2
- psB1C3 for Nir, 1M Betaine
- purification of psB1C3 USER fragment to make Biobricks (escaped from well -> Ethanol?)
- purification of HAO fragment (escaped from well -> Ethanol?)
- purification of AMO fragment (escaped from well -> Ethanol?)
- purification of psB1C3 USER fragment to make Biobricks (used more loading buffer)
- purification of HAO fragment (used more loading buffer)
- purification of AMO fragment (used more loading buffer)
- purification of pZA21::ara(tight)
- purification of pZA21::ara(tight) with endings for Nir
- 1 kb ladder
Conclusion
Navigate to the Previous or the Next Entry
 "
"