Team:Leeds/Project
From 2013.igem.org
m |
m |
||
| Line 43: | Line 43: | ||
<ul> | <ul> | ||
<li>[[File:Leeds_Microbeagleheader.png|700px|center|Beagles vs MicroBeagles|frameless]]</li> | <li>[[File:Leeds_Microbeagleheader.png|700px|center|Beagles vs MicroBeagles|frameless]]</li> | ||
| - | <li>[[File:Leeds_bb1schematic.png|700px|center|Our first | + | <li>[[File:Leeds_bb1schematic.png|700px|center|link=|Our first biosystem will be used to help characterise the Cpx pathway, which reacts to membrane stress|frameless]]</li> |
| - | <li>[[File:Leeds_Device2schematic.png|700px|center|Our second | + | <li>[[File:Leeds_Device2schematic.png|700px|center|link=|Our second biosystem will test the INP protein's capability to express any protein we like on the surface, in this case, a silica binding protein|frameless]]</li> |
| - | <li>[[File:Leeds_Device3chematic.png|600px|center|Finally, we will combine | + | <li>[[File:Leeds_Device3chematic.png|600px|center|link=|Finally, we will combine Biosystems 1 and 2, and make use of quorum sensing to produce a culture of super-sensitive micro-Beagles!|frameless]]</li> |
</ul> | </ul> | ||
</div></div> | </div></div> | ||
| Line 55: | Line 55: | ||
<br> | <br> | ||
==Phase I== | ==Phase I== | ||
| - | We split our project into self-contained phases, to help better organise ourselves, but also lay out a road map for our own future work within and without iGEM. The first of these looks at developing, characterising and producing basic | + | We split our project into self-contained phases, to help better organise ourselves, but also lay out a road map for our own future work within and without iGEM. The first of these looks at developing, characterising and producing basic Biosystems. |
| - | === | + | ===Biosystem 1=== |
| - | [[File:Leeds_bb1schematic.png|left|300px|Cartoon schematic of | + | [[File:Leeds_bb1schematic.png|left|300px|Cartoon schematic of Biosystem 1|link=|frameless]] |
| - | + | Biosystem 1 is a very simple modification of the Cpx promoter in ''E. coli'' to produce a fluorescent reporter when the cell is under membrane stress. We will then characterise and test the insertion following the work by [http://www.ncbi.nlm.nih.gov/pubmed/11830644 ''Otto & Silhaevy 2001''] in which glass beads were used to induce hydrophobic membrane stress. What we would hope is that when we add our cells to a vial containing similar hydrophobic beads we can detect fluorescence emissions confirming that our biobrick is working as we had hoped. Bio bricks we will be using: | |
BBa_K135000 pCpxR promoter responds to membrane stress | BBa_K135000 pCpxR promoter responds to membrane stress | ||
BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator | BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator | ||
<br style="clear:both"/> | <br style="clear:both"/> | ||
| - | === | + | ===Biosystem 2=== |
| - | [[File:Leeds_Device2schematic.png|right|300px|Cartoon schematic of | + | [[File:Leeds_Device2schematic.png|right|300px|Cartoon schematic of Biosystem 2|link=|frameless]] |
Simultaneously, we will be developing a BioBrick for physical attachment and detection of particles. This will work using Ice Nucleation Protein to display a oligo-peptide of our choice on the outer-membrane of our ''E. coli'', initially this will be a peptide capable of binding silica beads allowing us to create a model system of particle detection. The cells producing this receptor complex will also be constitutively producing a fluorescent reporter of our choice to allow for its detection on silica beads following removal of supernatant and washing. Biobricks we will be using: | Simultaneously, we will be developing a BioBrick for physical attachment and detection of particles. This will work using Ice Nucleation Protein to display a oligo-peptide of our choice on the outer-membrane of our ''E. coli'', initially this will be a peptide capable of binding silica beads allowing us to create a model system of particle detection. The cells producing this receptor complex will also be constitutively producing a fluorescent reporter of our choice to allow for its detection on silica beads following removal of supernatant and washing. Biobricks we will be using: | ||
BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator | BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator | ||
| Line 78: | Line 78: | ||
==Phase II== | ==Phase II== | ||
Phase II takes the products of Phase I to the next level, and begins to look at integration of the two BioBricks. | Phase II takes the products of Phase I to the next level, and begins to look at integration of the two BioBricks. | ||
| - | === | + | ===Biosystem 3=== |
| - | [[File:Leeds_Device3chematic.png|left|400px|Cartoon schematic of | + | [[File:Leeds_Device3chematic.png|left|400px|Cartoon schematic of Biosystem 3|link=|frameless]] |
| - | Due to it's size, the team have decided to collaborate with the [[Team:Evry|Evry]] team and make use of the Goldengate/GoldenBrick assembly methods. This will hopefully allow our small team to rather rapidly integrate and produce this | + | Due to it's size, the team have decided to collaborate with the [[Team:Evry|Evry]] team and make use of the Goldengate/GoldenBrick assembly methods. This will hopefully allow our small team to rather rapidly integrate and produce this third master Biosystem. |
<br style="clear:both"/> | <br style="clear:both"/> | ||
==The Cpx Pathway== | ==The Cpx Pathway== | ||
This project is highly dependent upon a naturally occuring pathway in ''E.Coli'' called the Cpx pathway. It is associated with the regulation of periplasmic membrane stress and the misfolding of surface proteins. We may need to fine tune MicroBeagle for different applications, so by understanding the way the pathway is regulated, we stand a better chance of controlling the exact response we want. This may be done in a number of ways, from utilisation of an off-switch regulator, CpxP, to additonal pathways used to create a bio-logic gate or even by optimisation of buffer solution. | This project is highly dependent upon a naturally occuring pathway in ''E.Coli'' called the Cpx pathway. It is associated with the regulation of periplasmic membrane stress and the misfolding of surface proteins. We may need to fine tune MicroBeagle for different applications, so by understanding the way the pathway is regulated, we stand a better chance of controlling the exact response we want. This may be done in a number of ways, from utilisation of an off-switch regulator, CpxP, to additonal pathways used to create a bio-logic gate or even by optimisation of buffer solution. | ||
}} | }} | ||
Revision as of 11:37, 3 September 2013
|
Our ProjectWhat is MicroBeagle I hear you cry?
Phase IWe split our project into self-contained phases, to help better organise ourselves, but also lay out a road map for our own future work within and without iGEM. The first of these looks at developing, characterising and producing basic Biosystems. Biosystem 1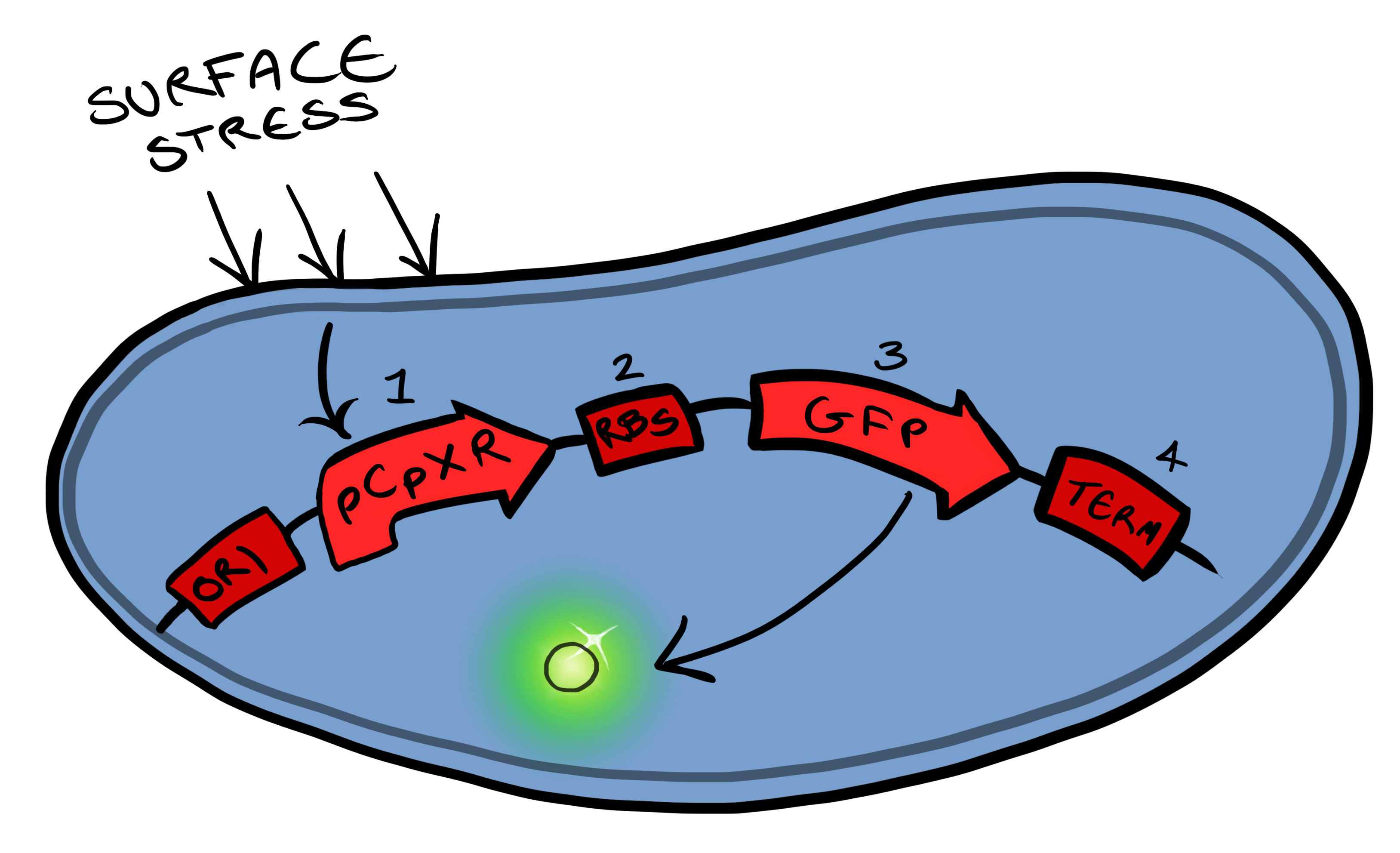 Biosystem 1 is a very simple modification of the Cpx promoter in E. coli to produce a fluorescent reporter when the cell is under membrane stress. We will then characterise and test the insertion following the work by [http://www.ncbi.nlm.nih.gov/pubmed/11830644 Otto & Silhaevy 2001] in which glass beads were used to induce hydrophobic membrane stress. What we would hope is that when we add our cells to a vial containing similar hydrophobic beads we can detect fluorescence emissions confirming that our biobrick is working as we had hoped. Bio bricks we will be using:
BBa_K135000 pCpxR promoter responds to membrane stress
BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator
Biosystem 2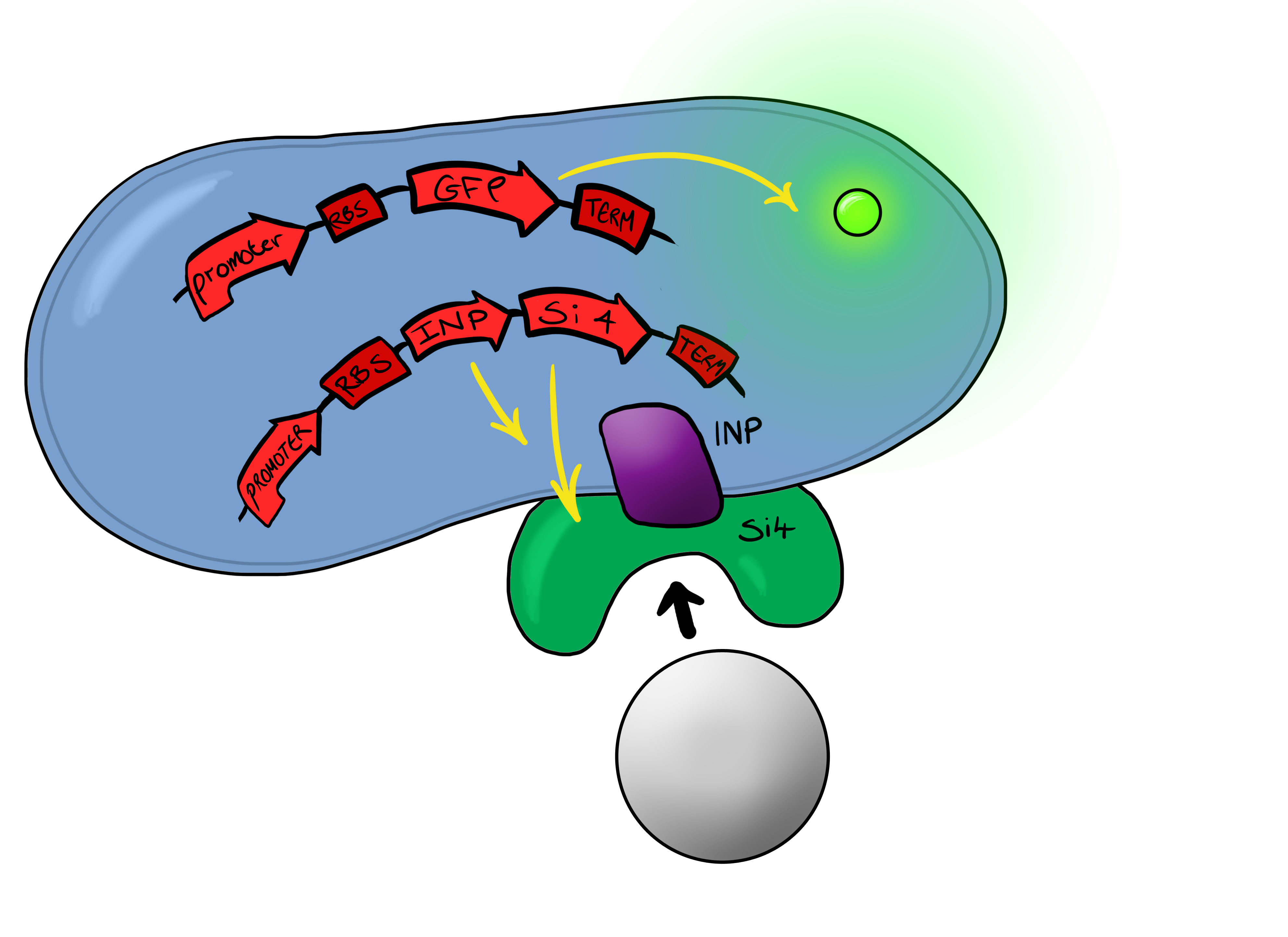 Simultaneously, we will be developing a BioBrick for physical attachment and detection of particles. This will work using Ice Nucleation Protein to display a oligo-peptide of our choice on the outer-membrane of our E. coli, initially this will be a peptide capable of binding silica beads allowing us to create a model system of particle detection. The cells producing this receptor complex will also be constitutively producing a fluorescent reporter of our choice to allow for its detection on silica beads following removal of supernatant and washing. Biobricks we will be using: BBa_K081012 Green fluorescent protein generator. Contain ribosome binding site and terminator BBa_K523008 Ice nucleation protein. Transported to cell membrane shows C-terminus on cell surface BBaa_B0015 Double terminator BBa_B0034 Strong ribosome binding site BBa_J23119 Constitutive promoter Again, fluorescent reporters will be used, but this time the focus is on the adhesion proteins being used.
Youtube VideosWhile doing all of this, the team aims to create short [http://www.youtube.com/user/igemleeds/videos Youtube] videos to teach Synthetic Biology at a range of levels, but especially including instructional videos for future team members.
Phase IIPhase II takes the products of Phase I to the next level, and begins to look at integration of the two BioBricks. Biosystem 3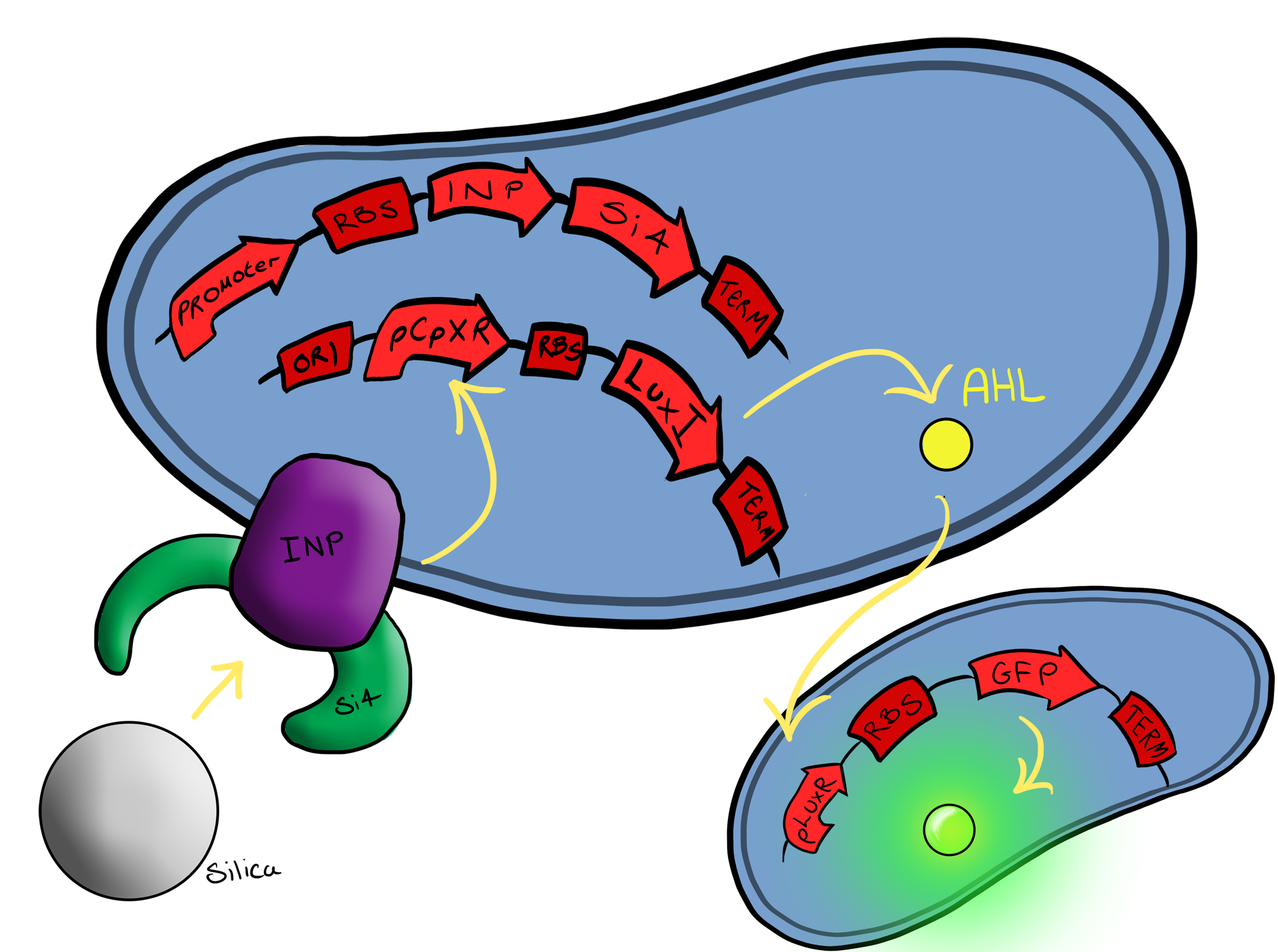 Due to it's size, the team have decided to collaborate with the Evry team and make use of the Goldengate/GoldenBrick assembly methods. This will hopefully allow our small team to rather rapidly integrate and produce this third master Biosystem.
The Cpx PathwayThis project is highly dependent upon a naturally occuring pathway in E.Coli called the Cpx pathway. It is associated with the regulation of periplasmic membrane stress and the misfolding of surface proteins. We may need to fine tune MicroBeagle for different applications, so by understanding the way the pathway is regulated, we stand a better chance of controlling the exact response we want. This may be done in a number of ways, from utilisation of an off-switch regulator, CpxP, to additonal pathways used to create a bio-logic gate or even by optimisation of buffer solution. | |||||||
 |
| ||||||

| |||||||

| |||||||
 "
"