Team:Newcastle/Project/plants
From 2013.igem.org
| Line 11: | Line 11: | ||
https://static.igem.org/mediawiki/2013/2/25/BareCillus_Plant_infographic.png | https://static.igem.org/mediawiki/2013/2/25/BareCillus_Plant_infographic.png | ||
| - | == | + | ==Benefits of using L-forms== |
<p>One component of our project is focused on introducing L-forms into plants. We have shown that they survive in, but not outside species such as Chinese cabbage. </p> | <p>One component of our project is focused on introducing L-forms into plants. We have shown that they survive in, but not outside species such as Chinese cabbage. </p> | ||
Revision as of 12:57, 20 September 2013

Contents |
L-forms in Plants
Overview
L-forms and plants can exist in a symbiotic relationship as plants provide an osmotically suitable environment. In return, L-forms can confer benefits to their host including reducing the rate of fungal infection. We washed chinese cabbage seedlings in a solution of green fluorescent protein (GFP) labelled L-forms, allowing the plants to take up the bacteria. We will then viewed our L-forms inside the plant using light and confocal microscopy.
In the future, L-forms could be engineered to supply nutrients to plants, potentially increasing crop yield in low fertility soil. Potential applications include L-forms which produce anti-fungal compounds could protect their host from infection and L-forms which allow nitrogen fixation in crops without root nodules. There is evidence that L-forms may [http://www.ncbi.nlm.nih.gov/pubmed/11849491 inhibit fungal pathogenesis in plants]. L-forms are osmotically sensitive, giving the ethical advantage that they lyse if they escape from their host plant into the environment.

Benefits of using L-forms
One component of our project is focused on introducing L-forms into plants. We have shown that they survive in, but not outside species such as Chinese cabbage.
L-forms could be modified to produce a benefit to the plant, producing nutrients or protection. Research has already shown prevention of fungal pathogenesis in Chinese cabbage and strawberry plants which have been exposed to B. subtilis L-forms, though the mechanism by which this works is not yet understood.
This may appear an unnecessary step- why not just modify the plant cells? First, bacteria are significantly easier to manipulate and genetically modify than plant cells, which are very complex. There are also bacteria genes, beneficial to plants, which would not be functional if transformed into a GM plant.
There are ethical boundaries associated with modifying plants. GM plants could well outcompete native species, spreading away from farmland and into the wild, becoming weeds and reducing biodiversity. They could also crossbreed with other strains.
Using non-L-form bacteria also has multiple potential pitfalls. Kill switches are never 100% safe as there is always the risk of mutations that turn off these genes, resulting in bacteria able to proliferate freely. L-forms however are incredibly osmotically sensitive, unable to survive outside of plants. Therefore they could be introduced into the environment- if they do leave the plants, they will very quickly pop due to water entering through osmosis, and a lack of a cell wall to counteract the resulting increased internal pressure. We have shown this experimentally using water extracted from soil.
Infecting plants with L-forms is therefore safer than infecting with normal bacteria and also safer than directly modifying plants
Methods
In order to prove that L-forms can grow in plants we soaked germinating chinese cabbage seedlings and soaked in a solution of GFP labelled L-forms. We then replanted the seedlings and viewed our L-forms via light microscopy before using confocal microscopy to generate better quality images. Our protocol is modified from [http://www.ncbi.nlm.nih.gov/pubmed/12859751 Tsomlexoglou et al.]:
Inoculating L-forms into Chinese Cabbage
- Rinse seeds for 2 minutes in 70% (v/v) ethanol.
- Soak seeds for 10 minutes in 20% (v/v) Milton’s sterilising fluid.
- Wash seeds thoroughly five times in sterile distilled water and leave in final wash for fifteen minutes.
- Place seeds in (9cm) Petri dishes (20-25 seeds per dish) containing [http://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Product_Information_Sheet/1/m5519pis.pdf Murashige and Skoog (M and S) basal medium], solidified with 0.8% (w/v) agar no.1 (Oxoid, UK) and incubate at 25 degrees Celsius in the dark until radicals just appear (this should take 21-24hr for Chinese cabbage).
- Harvest L-forms from 3-4 day old LPM plates using 5% (w/v) mannitol into sterile plastic universal tubes.
- Use spectrophotometry to produce a bacterial suspension containing approximately 107 CFU ml-1 (OD600: approx. 0.7).
- Select seeds with radicals 1-2mm in length and soak in the bacterial suspension (20 seeds per 10ml) for 3 hours at 25 degrees Celsius, gently shaking the seeds by hand every 30 minutes. Treat some seedlings with 5% (w/v) mannitol instead of L-forms to act as a control.
- Wash seeds ten times in distilled water.
- Replant plants on M and S agar plates.
- Incubate seeds in sunlight for between 24 hours and 7 days at 25°C.
- The plants are ready to be viewed using microscopy.
The washing of seeds in ethanol and Milton’s sterilising fluid is carried out to kill any bacteria residing on the surface of the seed. M and S medium is a growth media designed for the cultivation of plants. Washing the seeds in distilled water after soaking them in bacterial solution lyses any L-forms not inside the plant. We carried out our microscopy 4 days after the seeds were soaked in L-forms, to co-inside with when [http://www.ncbi.nlm.nih.gov/pubmed/12859751 Tsomlexoglou et al.] carried out PCR to prove the presence of L-forms in their chinese cabbage.
Alternative Methods Considered
Before deciding to use Green Fluorescent Protein labelled L-forms, we considered a couple of alternative ways to detect L-forms in plants:
β-glucuronidase assay
In order to prove that L-forms can grow in plants, we considered transforming B. subtillis strain LR2 with the gusA reporter gene coding for β-glucuronidase. We would have removed gusA BioBrick from its plasmid backbone and attach it to another plasmid containing AprE for homologous recombination and kanamycin resistance for selection. We would then have transformed B. subtillis with the new transformation vector. The bacteria that have taken up the plasmid can would have been selected for via kanamycin resistance. A transformation vector would have been used rather than a normal plasmid so that gusA would integrate through homologous recombination into the bacteria’s chromosome, securing the expressing of the gusA.
The β-glucuronidase produced by the L-forms in plants would cleave the colourless substrate 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-Gluc) to produce a blue product allowing the L-form distribution in plant tissue to be observed. We considered this method after reading a paper by [http://www.ncbi.nlm.nih.gov/pubmed/12859751 Tsomlexoglou et al.]
Enzyme-linked immunosorbent assay (ELISA)
We considered producing an ELISA to detect L-forms. We could have generated antibodies against a L-form specific antigen. Alternatively we considered introducing a novel gene into L-forms to produce a novel antigen to generate antibodies against. However this would have involved using animals to generate polyclonal antibodies. We decided this raised ethical issues and was too time consuming to complete in our ten week placement. We considered this method after reading a paper by [http://www.ncbi.nlm.nih.gov/pubmed/11069643 Ferguson et al.]
DAPI Staining
[http://probes.invitrogen.com/media/pis/mp01306.pdf 4',6-diamidino-2-phenylindole (DAPI)] can be used to stain both bacterial and plant DNA fluorescent blue. For this reason we considered using DAPI to counter-stain the plant genome and inoculating the seedlings with GFP-labelled L-forms so we can see both the plant nucleus and our L-forms. Alternatively we could have stained GFP-labelled L-forms with DAPI so that when viewing L-forms with confocal microscopy we could check if they fluoresce blue as well as green. This would add further evidence that we had successfully inoculated L-forms into chinese cabbage.
PCR
[http://www.ncbi.nlm.nih.gov/pubmed/12859751 Tsomlexoglou et al.] used the polymerase chain reaction (PCR) to detect L-forms containing the gusA gene in four day old chinese cabbage. Primers specific to the gusA gene were used as this gene was specific to the genetically modified L-forms. We considered using primes specific to our GFP-labelled L-forms to carry out PCR. We chose not to carry out this method due to time constraints and we wanted a more aesthetic detection method.
Results
Results Log
09/08/13
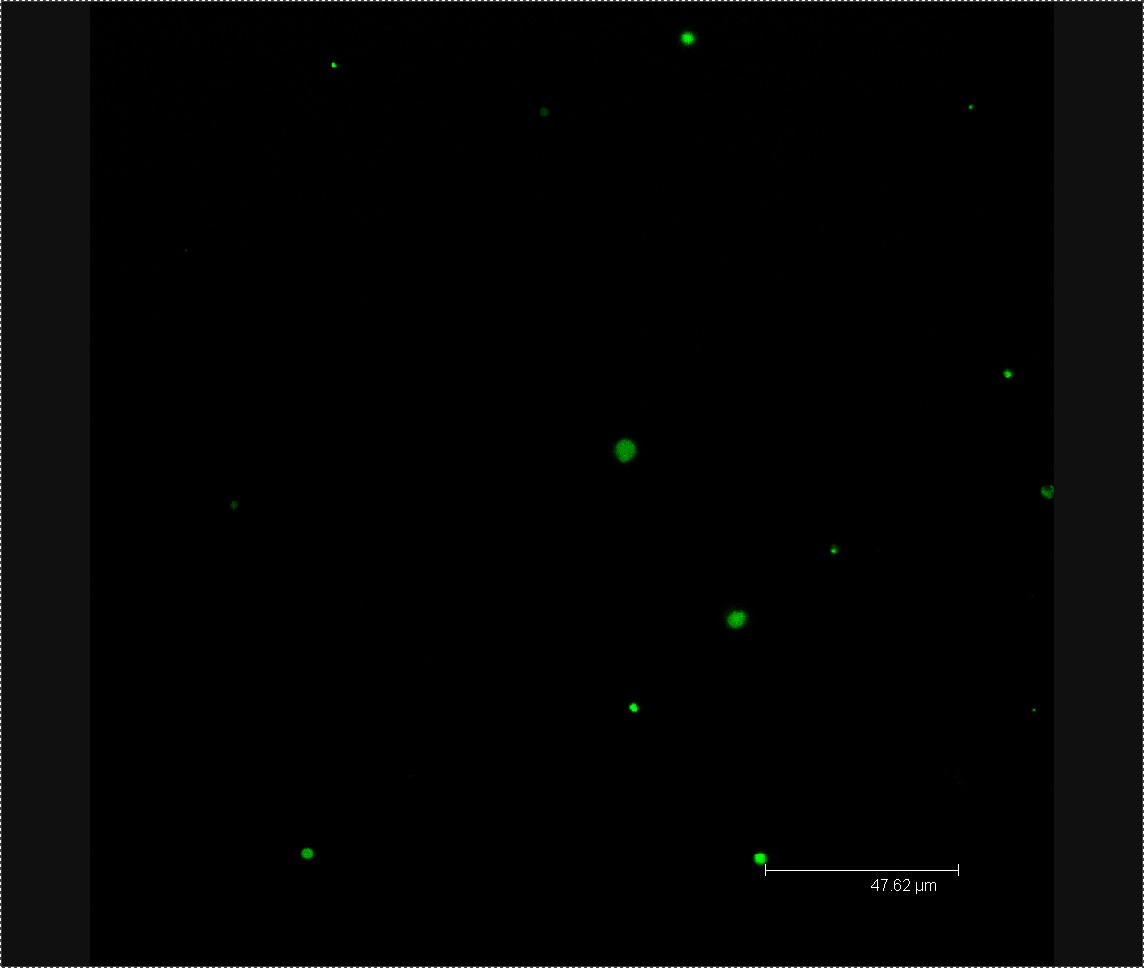
Today we viewed the seedlings, that we had washed with L-forms yesterday,using bright field and fluorescence microscopy. When viewing an L-form washed seedling using fluorescence microscopy we noticed clearly defined green circles in several different areas of the plant. These were of the correct size (about 0.5um) to correspond to our GFP labelled L-forms. No such green circles were found in our control that had been washed in mannitol solution instead of L-form solution. These results are encouraging as they indicate we may have successfully inoculated plants with L-forms, although more evidence is needed.
14/08/13
Today we viewed our plants using confocal microscopy to try and detect any GFP-labelled bacteria. After viewing a few seeds washed in L-form solution under the microscope, we found a green fluorescing dot in the correct size range to be an L-form. No green dots were found in the rod cell or mannitol controls. Following these results we have decided to use confocal microscopy four days after washing our plants in L-form solution, rather than after one. This co-insides with when [http://www.ncbi.nlm.nih.gov/pubmed/12859751 Tsomlexoglou et al.] used detected L-forms in their chinese cabbage using PCR.
Figure 1. Possible L-form inside Chinese cabbage. The bright green dot is the correct size to be an L-form. The green fluorescence image used to detect the L-form is superimposed onto a bright-field image of plant matter.
Figure 2. L-forms from the solution used to wash the Chinese cabbage.
23/08/13
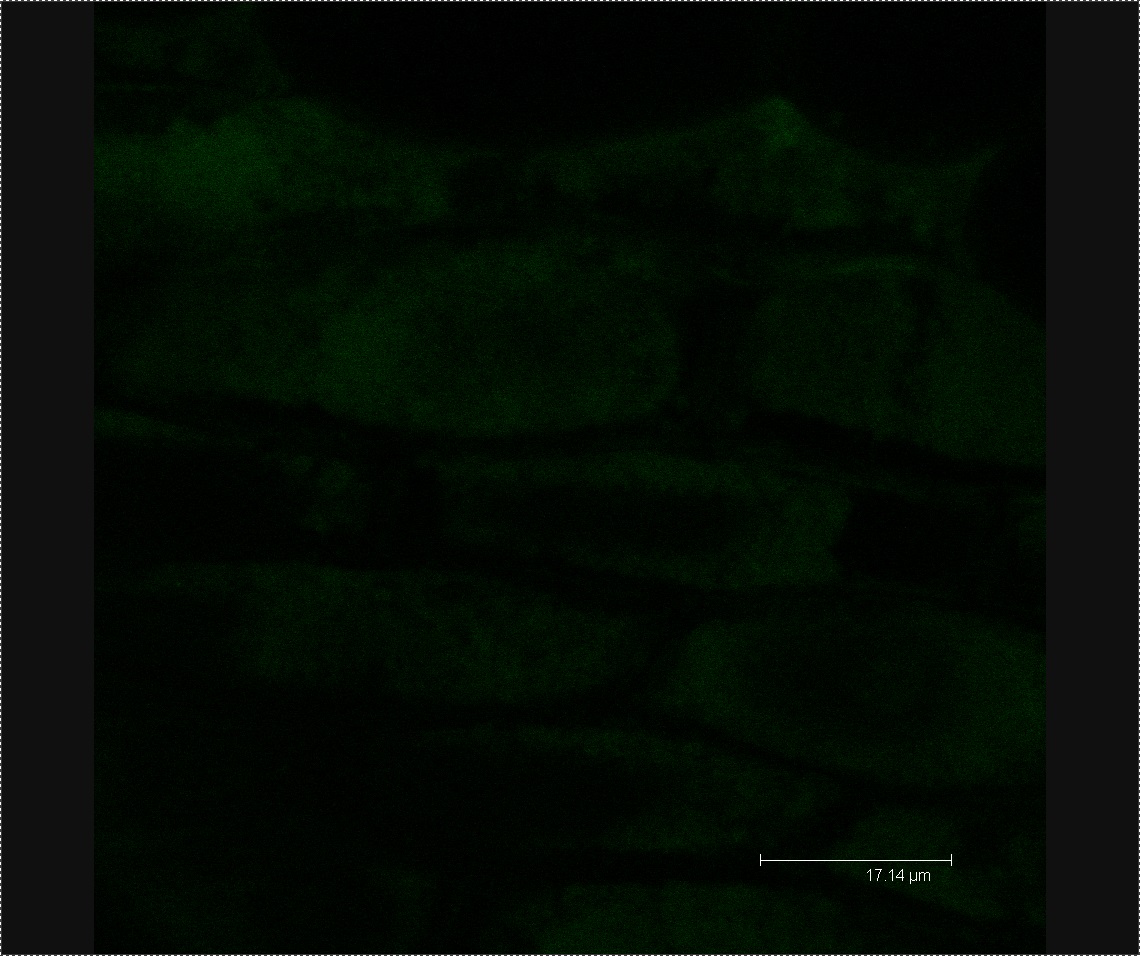
We viewed 7 different Chinese cabbage seedlings washed with L-forms under the confocal microscope. In three of these plants we found clusters of green fluorescent dots about 0.5µm in diameter. We found no clusters of green fluorescent dots in three seedlings soaked in the mannitol control.
Figure 3. Cluster of L-forms inside a Chinese cabbage seedling.
Figure 4. Image of mannitol-soaked control exhibiting faint green auto-fluorescence, allowing us to see the outline of plant cells.
The Future
Nitrogen Fixation
References
[http://www.ncbi.nlm.nih.gov/pubmed/11069643 Ferguson, C.M.J., Booth, N.A. and Allan, E.J. (2000) An ELISA for the detection of Bacillus subtilis L-form bacteria confirms their symbiosis in strawberry. Letters in Applied Microbiology. 31(5), p. 390-394.]
[http://probes.invitrogen.com/media/pis/mp01306.pdf INVITROGEN. (2006) DAPI Nucleic Acid Stain. (online) Available from: http://probes.invitrogen.com/media/pis/mp01306.pdf (Accessed 28 August 2013)]
[http://www.ncbi.nlm.nih.gov/pubmed/12859751 Tsomlexoglou, E., Daulagala, P.W.H.K.P., Gooday, G.W., Glover, L.A., Seddon, B. and Allan, E.J. (2003) Molecular detection and β-glucuronidase expression of gus-marked Bacillus subtilis L-form bacteria in developing Chinese cabbage seedlings. Journal of Applied Microbiology. 95(2), p. 218-224.]
[http://www.ncbi.nlm.nih.gov/pubmed/11849491 Walker, R. , Ferguson, C.M.J. , Booth, N.A. and Allan, E.J. (2002) The symbiosis of Bacillus subtilis L-forms with Chinese cabbage seedlings inhibits conidial germination of Botrytis cinerea. Letters in Applied Microbiology. (34) p.42–45.]
 "
"