Template:Kyoto/Notebook/Sep 2
From 2013.igem.org
(Difference between revisions)
(→Gel Extraction) |
(→Restriction Enzyme Digestion) |
||
| Line 75: | Line 75: | ||
|NC(X+P)||0.5µL||0µL||0µL||0µL||0µL||0µL||1µL||0.1µL||8.4µL||10µL | |NC(X+P)||0.5µL||0µL||0µL||0µL||0µL||0µL||1µL||0.1µL||8.4µL||10µL | ||
|} | |} | ||
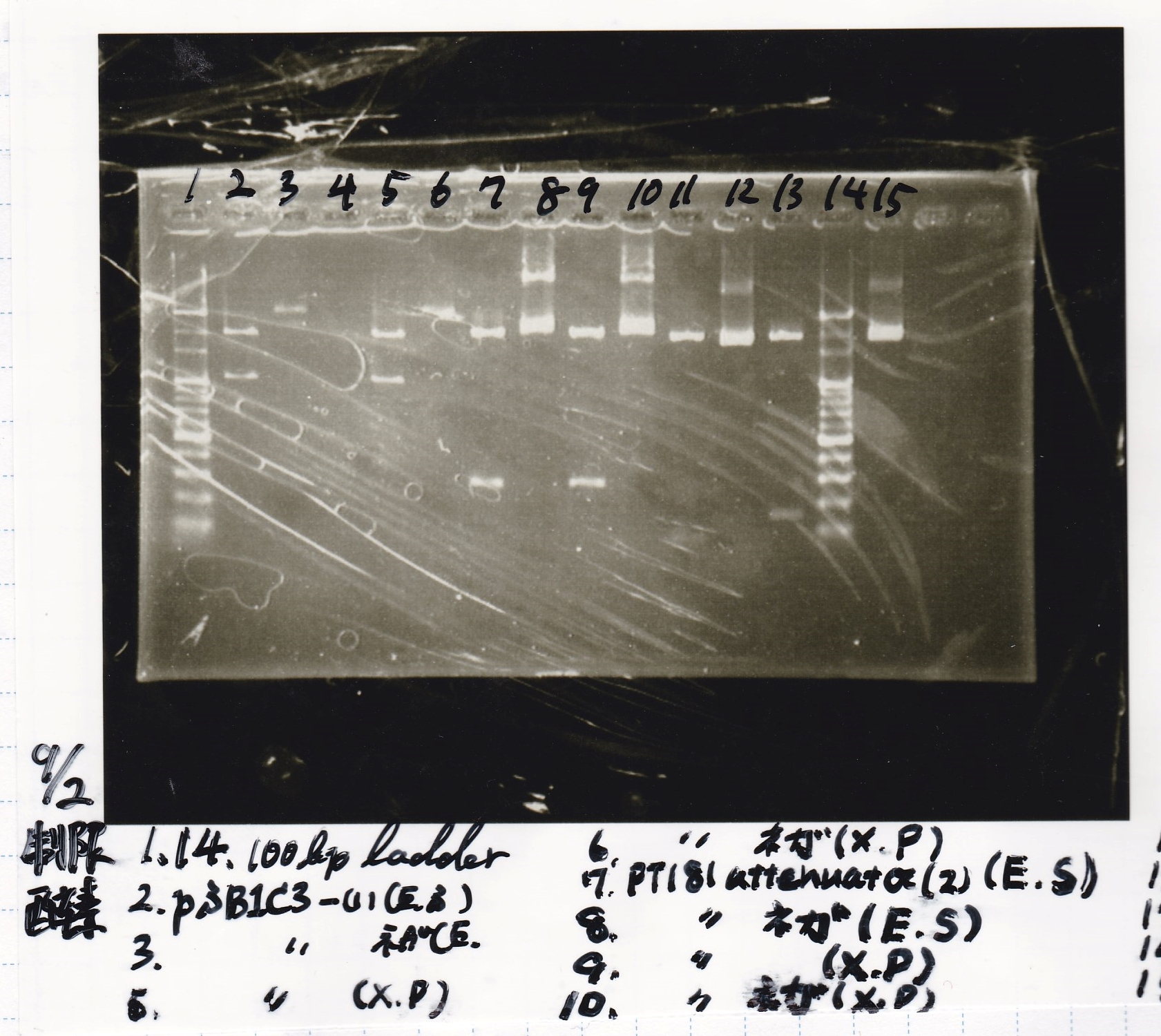
| + | [[File:Igku Sep2 Restriction Enzyme Digestion (N1-1).jpg]]<br> | ||
</div> | </div> | ||
Revision as of 12:53, 24 September 2013
Contents |
Sep 2
Liquid Culture
| Sample | medium |
|---|---|
| 9/1 entA-(Master Plate)-1 | 4mL SOB(+Km) |
37°C
Colony PCR
| Sample | base pair |
|---|---|
| 9/1 RBS-lysis2-DT-(1) | 985 |
| Ptet-RBS-lacZα-DT-(1) | 765 |
| Plac-RBS-lacZα-DT-(1) | 765 |
| Plac-RBS-lacZα-DT-(2) | 765 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 1min | 30cycles |
Restriction Enzyme Digestion
| pSB1C3-(1) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 cuts(E+S) | 7µL | 1µL | 1µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 17.7µL | 30µL |
| NC(E+S) | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.6µL | 10µL |
| 2 cut(X+P)s | 7µL | 0µL | 0µL | 1µL | 1µL | 0µL | 3µL | 0.3µL | 17.7µL | 30µL |
| NC(X+P) | 0.3µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.6µL | 10µL |
| 8/21 tRMA-spinach(1) | EcoRI | SpeI | BufferB | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 8µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 17.6µL | 30µL |
| NC | 0.5µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL |
| 8/21 tetR aptamer12_1R-(1) | EcoRI | SpeI | BufferB | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 6µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 18.7µL | 30µL |
| NC | 0.4µL | 0µL | 0µL | 1µL | 0.1µL | 8.5µL | 10µL |
| 8/21 pT181 attenuator-(2) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 cuts(E+S) | 8µL | 1µL | 1µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 16.3µL | 30µL |
| NC(E+S) | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.4µL | 10µL |
| 2 cut(X+P)s | 8µL | 0µL | 0µL | 1µL | 1µL | 0µL | 3µL | 0.3µL | 16.3µL | 30µL |
| NC(X+P) | 0.5µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL |
Liquid Culture
| Sample | medium | |||
|---|---|---|---|---|
| tRNA-Spinach-1 | Plusgrow medium(+CP) | |||
| tetR aptamaer12_1R-1 | Plusgrow medium(+CP) | |||
| pT181 attenuator-1 | Plusgrow medium(+CP) | = | pT181 antisense-1 | Plusgrow medium(+CP) |
Master Plate
| Number | Use LB plate(+CP) |
|---|---|
| 1 | tRNA Spinach-1 |
| 2 | tetR aptamaer12_1R |
| 3 | tetR aptamaer12_P |
| 4 | tetR aptamaer12_1M |
| 5 | pT181 attenuator-2 |
| 6 | Fusion1 attenuator-1 |
| 7 | Fusion3m2 attenuator-1 |
| 8 | pT181 antisense-1 |
| 9 | Fusion1 antisense-1 |
| 10 | Fuaion6 antisense-1 |
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | - |
| 2 | pSB1C3(E+S) | EcoRI&SpeI |
| 3 | ||
| 4 | -- | -- |
| 5 | 8/24 tetR aptamer 12_1R | EcoRI&SpeI |
| 6 |
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bp ladder | - |
| 2 | pT181 attenuator(2)(E+S) | EcoRI&SpeI |
| 3 | ||
| 4 | -- | -- |
| 5 | pT181 attenuator(2)(X+P) | EcoRI&SpeI |
| 6 | ||
| 7 | -- | -- |
| 8 | pT181 attenuator(2)(X+P) | EcoRI&SpeI |
| 9 | ||
| 10 | -- | -- |
| 11 | Spinach | EcoRI&SpaI |
| 12 |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| pSB1C3 (EcoRI&SpeI) | 3.0 | 2.18 | 0.31 |
| pSB1C3 (XbaI&PstI) | 4.7 | 2.25 | 0.36 |
| pT181 attenuator-(2)(EcoR&SpeI) | 8.6 | 2.74 | 0.01 |
| pT181 attenuator-(2) (XbaI&PstI) | 16.5 | 2.46 | 0.03 |
| Spinach (EcoRI&SpeI) | 2.8 | 2.98 | 0.27 |
| tetR aptamer12_1R (EcoRI&SpeI) | 50.4 | 28.07 | 1.97 |
Colony PCR
| Sample | base pair |
|---|---|
| 9/1 Spinach(pSB1C3) | about 450 |
| 9/1 Spinach-DT | 596 |
| 9/1 RBS-lysis1-DT | 613 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 40s | 30cycles |
File:Igku sep2electrophoresis1.png
| Sample | base pair |
|---|---|
| 9/1 Ptet-RBS-lacZα-DT-2 | 738 |
| 9/1 Ptet-RBS-lacZα-DT-3 | 738 |
| 9/1 Ptet-RBS-lacZα-DT-4 | 738 |
| 9/1 Ptet-RBS-lacZα-DT-5 | 738 |
| 9/1 Ptet-RBS-lacZα-DT-6 | 738 |
| 9/1 RBS-lysis2-DT-2 | 985 |
| 9/1 RBS-lysis2-DT-3 | 985 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 1min | 30cycles |
Master Plate
| Number | Use LB plate(+CP) |
|---|---|
| 1 | 9/1 Spinach(pSB1C3) |
| 2 | 9/1 Spinach-DT-(1) |
Liquid Culture
| Sample | medium |
|---|---|
| pT181 attenuator | Plusgrow medium (+CP) |
| pT181 antisense | Plusgrow medium (+CP) |
| Spinach | Plusgrow medium (+CP) |
- 37&dig:C ihour incubate
Transformation
| Name | Sample | Competent Cells | Total | Plate |
|---|---|---|---|---|
| 8/28 RBS-lysis3-DT | 3µL | 30µL | 33µL | CP |
 "
"