From 2013.igem.org
(Difference between revisions)
|
|
| Line 373: |
Line 373: |
| | |12||1kbp ladder||--||-- | | |12||1kbp ladder||--||-- |
| | |} | | |} |
| - | [[File:igku_xx.xxx]]<br> | + | [[File:Igku Sep5 Electrophoresis(N6) 3.jpg]]<br> |
| | </div> | | </div> |
| | | | |
Revision as of 13:17, 25 September 2013
Sep 5
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | J23100-3 | -- | --
|
| 3 | J23100-4 | -- | --
|
| 4 | J23100-5 | -- | --
|
| 5 | 100bp ladder | -- | --
|
File:Igku xx.xxx
Miniprep
Nakamoto
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 8/3 J23100-3 | 349.1 | 1.69 | 1.851
|
| 8/10 Plac-2 | 331.3 | 1.84 | 1.42
|
Ligation
Kojima
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | Pconst | 13.2 | spinach-DT | 2.1 | 3.5
|
| experiment | Plac | 15.3 | spinach-DT | 2.1 | 3.5
|
| experiment | Pconst | 13.2 | pT181 antisense (XbaI & PstI) | 6.6 | 3.5
|
| experiment | Plac | 15.3 | pT181 antisense (XbaI & PstI) | 6.6 | 3.5
|
| experiment | Ptet | 30.1 | pT181 antisense (XbaI & PstI) | 6.6 | 3.5
|
| experiment | Pconst | 13.2 | pT181 attenuator | 11.7 | 3.5
|
| experiment | Plac | 15.3 | pT181 attenuator | 11.7 | 3.5
|
| experiment | Ptet | 30.1 | RBS-lacZα-DT | 3.0 | 3.5
|
incubate 16 °C 1 hour
Electrophoresis
No name
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder |
|
| 2 | RBS-luxI-DT |
|
| 3 | RBS-luxI-DT |
|
| 4 | |
|
| 5 | Pbad/araC |
|
| 6 | Pbad/araC |
|


| Name | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-luxI-DT | 23.2 | 0.83 | 7.89
|
| Pbad/araC | 51.8 | 1.20 | 0.11
|
Colony PCR
No name
| Sample | base pair
|
| 9/4 aptamer 12_1R(pSB1C3)-1 | 384
|
| 9/4 aptamer 12_1R(pSB1C3)-2 | 384
|
| 9/4 pT181 antisense(pSB1C3)-1 | 415
|
| 9/4 pT181 antisense(pSB1C3)-2 | 415
|
| 9/4 pT181 antisense(pSB1C3)-3 | 415
|
| 9/4 pT181 antisense(pSB1C3)-4 | 415
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
| Sample | base pair
|
| 9/4 aptamer 12_1R-DT-1 | 521
|
| 9/4 aptamer 12_1R-DT-2 | 521
|
| 9/4 aptamer 12_1R-DT-3 | 521
|
| 9/4 aptamer 12_1R-DT-4 | 521
|
| 9/4 pT181 attenuator(pSB1C3) | 601
|
| 9/4 pT181 attenuator(pSB1C3) | 601
|
| 9/4 RBS-lysis1-DT | 613
|
| 9/4 RBS-lysis1-DT | 613
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 36s | 30cycles
|
| Sample | base pair
|
| 9/4 RBS-lysis2-DT-1 | 985
|
| 9/4 RBS-lysis2-DT-2 | 985
|
| 9/4 RBS-lysis2-DT-3 | 985
|
| 9/4 RBS-lysis2-DT-4 | 985
|
| 9/4 RBS-lysis3-DT-1 | 1210
|
| 9/4 RBS-lysis3-DT-2 | 1210
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 72s | 30cycles
|
Liquid Culture
Nakamoto
| Sample | medium
|
| 8/9 J23100-5 | Plusgrow medium(+Amp)
|
37°C
Transformation
No name
| Name | Sample(µL) | Competent Cells(µL) | Total(µL) | Plate
|
| 9/5 Pconst+spinach-DT | 2 | 20 | 22 | --
|
| 9/5 Plac+spinach-DT | 2 | 20 | 22 | --
|
| 9/5 Pconst+pT181 antisense | 2 | 20 | 22 | --
|
| 9/5 Plac+pT181 antisense | 2 | 20 | 22 | --
|
| 9/5 Ptet+pT181 antisense | 2 | 20 | 22 | --
|
| 9/5 Plux+pT181 attenuater | 2 | 20 | 22 | --
|
| 9/5 Ptet+pT181 attenuater | 2 | 20 | 22 | --
|
| 9/5 Ptet+pT181 attenuater | 2 | 20 | 22 | --
|
| 9/5 Pconst+RBS-tetR+DT | 2 | 20 | 22 | --
|
| 9/16(2012) T7-His-FT | 2 | 20 | 22 | --
|
| 9/16(2012) pBr322 | 2 | 20 | 22 | --
|
Restriction Enzyme Digestion
No name
| | 9/5 Pbad/araC | E | S | X | P | BSA | Buffer | MilliQ | total
|
| 2 cuts | 4µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 18µL | 30µL
|
| | 9/5 RBS-luxI-DT | E | S | X | P | BSA | Buffer | MilliQ | total
|
| 2 cuts | 4µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 18µL | 30µL
|
| | 8/28 Plux | E | S | X | P | BSA | Buffer | MilliQ | total
|
| 2 cuts | 12µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 10µL | 30µL
|
| NC | 0.6µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.4µL | 10µL
|
| | 9/5 Pconst(J23100) | E | S | X | P | BSA | Buffer | MilliQ | total
|
| 2 cuts | 5.7µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 16.3µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 9/5 Plac | E | S | X | P | BSA | Buffer | MilliQ | total
|
| 2 cuts | 6.0µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 16µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 8/31 Plux-RBS-GFP-DT-1 | E | S | X | P | BSA | Buffer | MilliQ | total
|
| 2 cuts | 9.4µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 12.6µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| | 8/20 Pconst-RBS-luxR-DT | E | S | X | P | BSA | Buffer | MilliQ | total
|
| 2 cuts | 4.5µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 13.5µL | 30µL
|
| NC | 0.2µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.8µL | 10µL
|
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/5 RBS-luxI-DT | 7.6 | 1.56 | -1.42
|
| 9/5 Pbad/araC | 7.5 | 1.54 | 3.84
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | aptamer 12_1R (pSB1C3)-1 | -- | --
|
| 3 | aptamer 12_1R (pSB1C3)-2 | -- | --
|
| 4 | pT181 antisense(pSB1C3)-1 | -- | --
|
| 5 | pT181 antisense(pSB1C3)-2 | -- | --
|
| 6 | pT181 antisense(pSB1C3)-3 | -- | --
|
| 7 | pT181 antisense(pSB1C3)-4 | -- | --
|
| 8 | 100bp ladder | -- | --
|
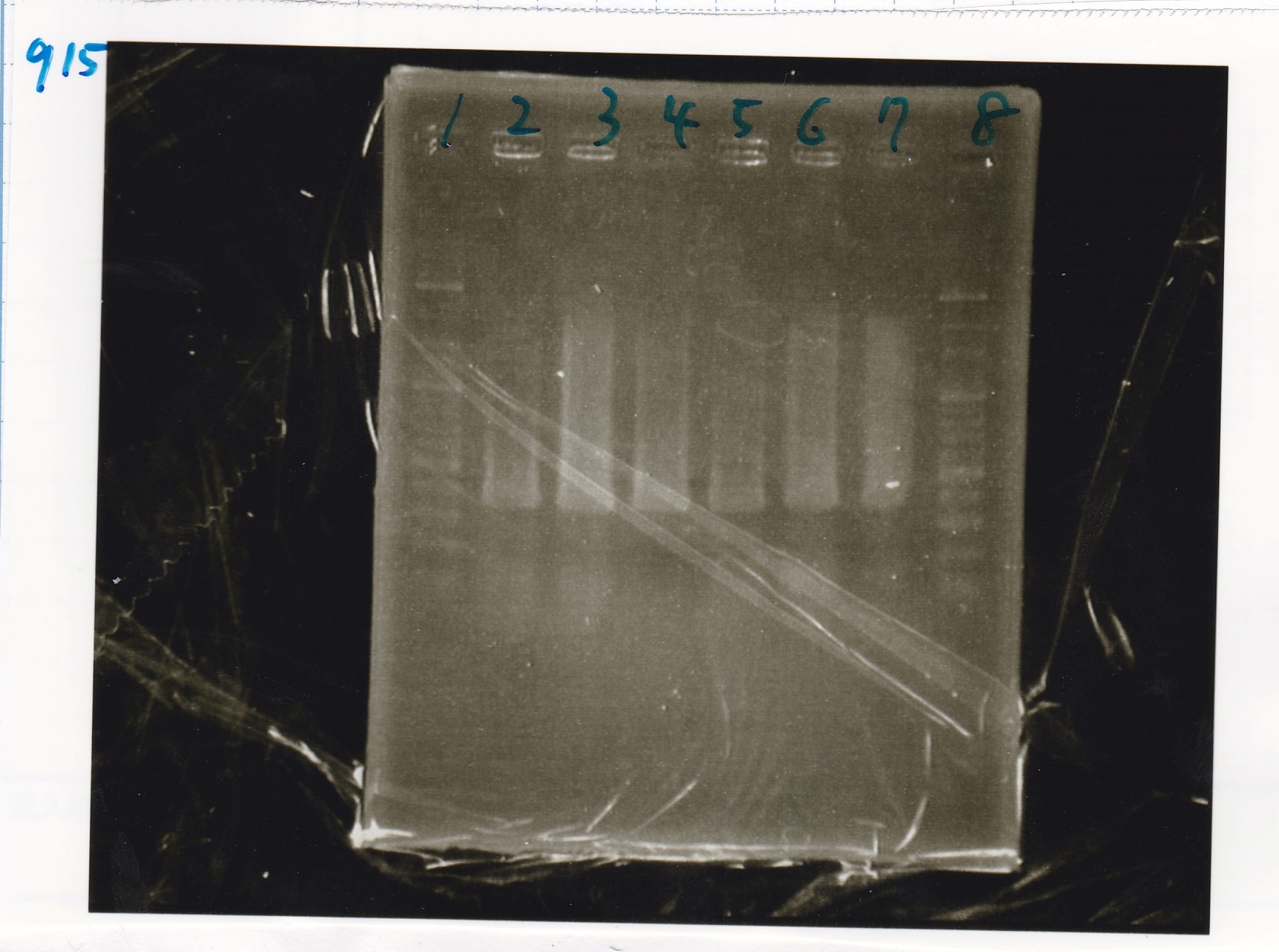
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | aptamer 12_1R-DT-1 | -- | --
|
| 3 | aptamer 12_1R-DT-2 | -- | --
|
| 4 | aptamer 12_1R-DT-3 | -- | --
|
| 5 | aptamer 12_1R-DT-4 | -- | --
|
| 6 | pT181 attenuator(pSB1C3)-1 | -- | --
|
| 7 | pT181 attenuator(pSB1C3)-2 | -- | --
|
| 8 | RBS-lysis1-DT-1 | -- | --
|
| 9 | RBS-lysis1-DT-2 | -- | --
|
| 10 | 100bp ladder | -- | --
|
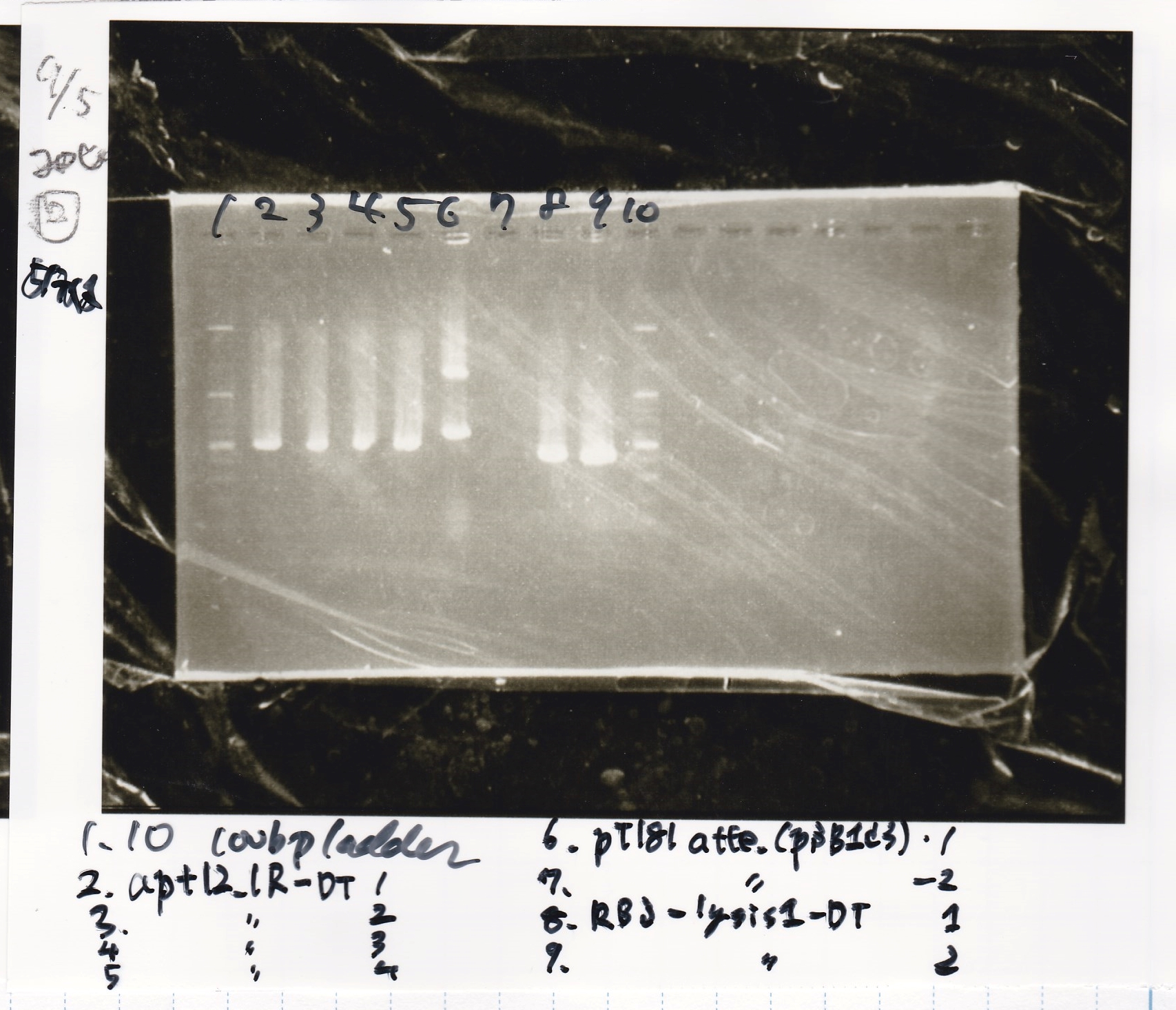
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | RBS-lysis2-DT-1 | -- | --
|
| 3 | RBS-lysis2-DT-2 | -- | --
|
| 4 | RBS-lysis2-DT-3 | -- | --
|
| 5 | RBS-lysis2-DT-4 | -- | --
|
| 6 | RBS-lysis3-DT-1 | -- | --
|
| 7 | RBS-lysis3-DT-2 | -- | --
|
| 8 | 100bp ladder | -- | --
|
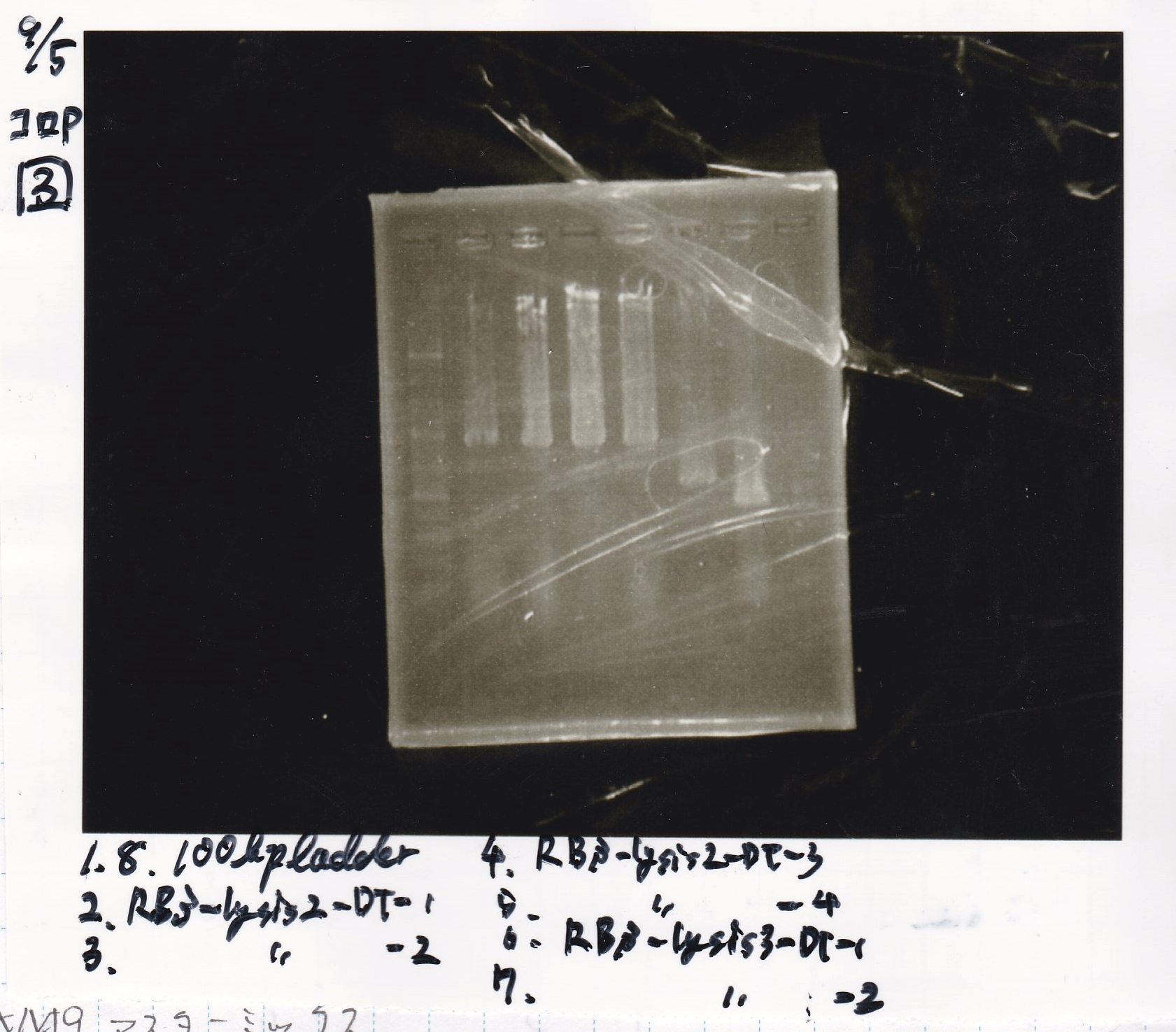
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kbp ladder | -- | --
|
| 2 | Pconst | SpeI | PstI
|
| 3 | Pconst NC | -- | --
|
| 4 | Plac | SpeI | PstI
|
| 5 | Plac NC | -- | --
|
| 6 | Plux | SpeI | PstI
|
| 7 | Plux NC | -- | --
|
| 8 | Pconst-RBS-luxR-DT | EcoRI | XbaI
|
| 9 | Pconst-RBS-luxR-DT NC | -- | --
|
| 10 | Plux-RBS-GFP-DT | EcoRI | SpeI
|
| 11 | Plux-RBS-GFP-DT NC | -- | --
|
| 12 | 1kbp ladder | -- | --
|
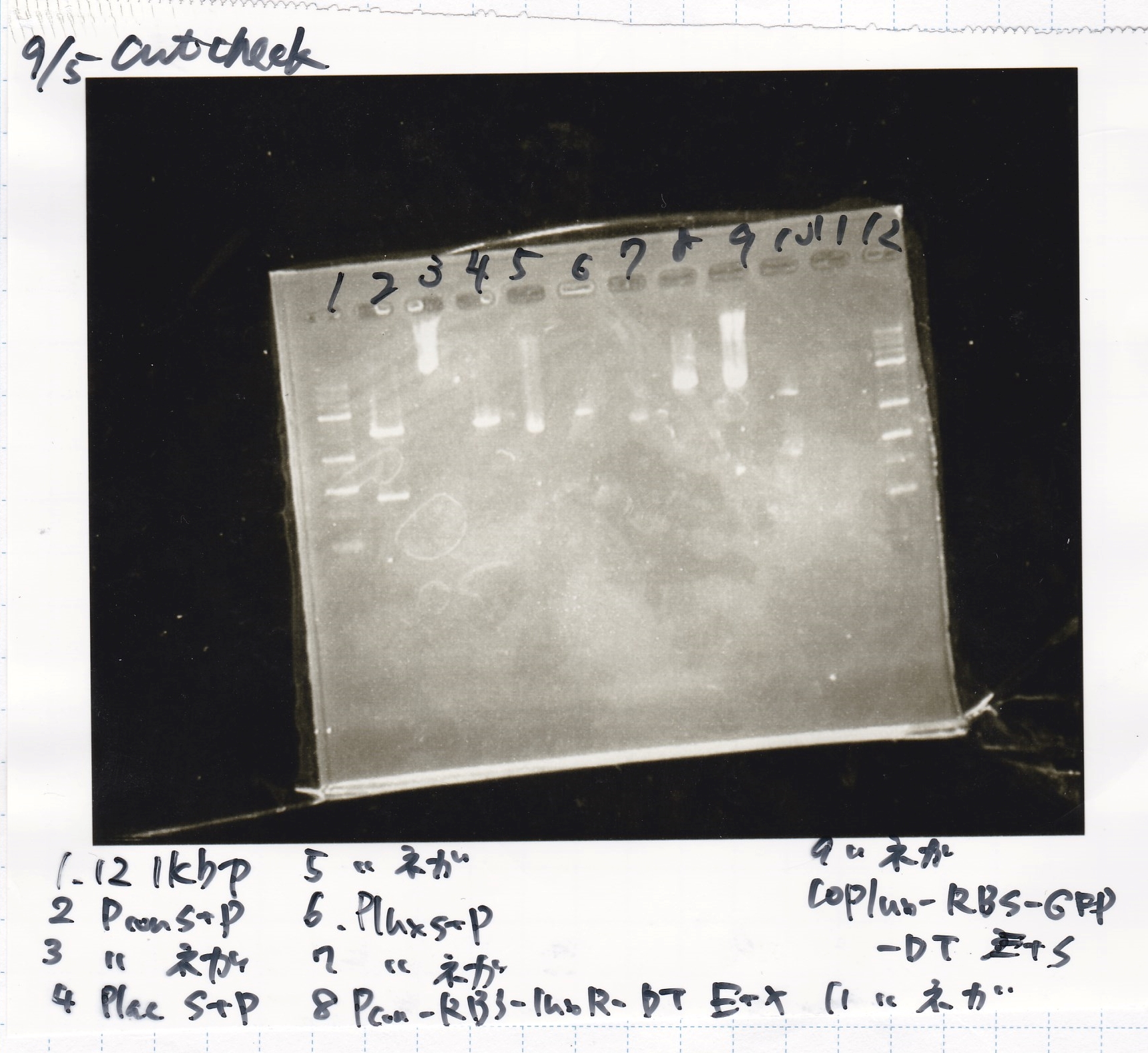
5x M9 Medium (+EDTA)
Hirano
| volume | 10ml
|
| Na2HPO4 | 60mg
|
| KH2PO4 | 30mg
|
| NaCl | 5mg
|
| NH4Cl | 10mg
|
| Fe(III)-EDTA | 1263.27mg
|
| MilliQ | up to 10 mL
|
- autoclave at 121 °C for 20 min
No name
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder |
|
| 2 | Pconst | SpeI & PstI
|
| 3 | Pconst | SpeI & PstI
|
File:Igku xxbeforexx.xxx
File:Igku xxafterxx.xxx
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pconst | 5.3 | 1.98 | 0.06
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 2 | Plac | SpeI & PstI
|
| 3 | Plac | SpeI & PstI
|
| 5 | Plux | SpeI & PstI
|
| 6 | Plux | SpeI & PstI
|
File:Igku xxbeforexx.xxx
File:Igku xxafterxx.xxx
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Plac (SpeI & PstI) | 5.2 | 1.82 | 0.36
|
| Plux(SpeI & PstI) | 8.0 | 1.87 | 0.25
|
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder |
|
| 2 | Pconst-RBS-luxR-DT | EcoRI & XbaI
|
| 3 | Pconst-RBS-luxR-DT | EcoRI & XbaI
|
| 5 | Plux-RBS-GFP-DT | EcoRI & SpeI
|
| 6 | Plux-RBS-GFP-DT | EcoRI & SpeI
|
File:Igku xxbeforexx.xxx
File:Igku xxafterxx.xxx
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pconst-RBS-luxR-DT(EcoRI & XbaI) | 30.6 | 1.84 | 1.16
|
| Plux-RBS-GFP-DT (EcoRI & SpeI) | 21.8 | 1.87 | 0.98
|
DNA Purification
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/5 RBS-luxI-DT | 7.6 | 1.56 | -1.42
|
| 9/5 Pbad/araC | 7.5 | 1.54 | 3.84
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 8/9 J23100-5 | 243.0 | 1.90 | 1.81
|
Colony PCR
No name
| Sample | base pair
|
| 9/4 RBS-lysis3-DT-3 | 1210
|
| 9/4 RBS-lysis3-DT-4 | 1210
|
| 9/4 RBS-lysis3-DT-5 | 1210
|
| 9/4 RBS-lysis1-DT-3 | 613
|
| 9/4 Ptrc KaiC-1 | --
|
| 9/4 pSB4K5 | 1370
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min 25s | 30cycles
|
Liquid Culture
Hirano
| Sample | medium
|
| 9/4 aptamer 12_1R-DT-1 | Plusgrow medium (+CP)
|
| 9/4 pT181 attenuator(pSB1C3)-1 | Plusgrow medium (+CP)
|
| 9/4 RBS-lysis2-DT-1 | Plusgrow medium (+CP)
|
| 9/4 Ptrc-KaiC -1 | Plusgrow medium (+Amp)
|
| 9/4 pSB4K5 -1 | Plusgrow medium (+Kan)
|
 "
"





