Team:UGA-Georgia/Plan
From 2013.igem.org
(→General Purpose and Goal) |
(→General Purpose and Goal) |
||
| Line 3: | Line 3: | ||
=General Purpose and Goal = | =General Purpose and Goal = | ||
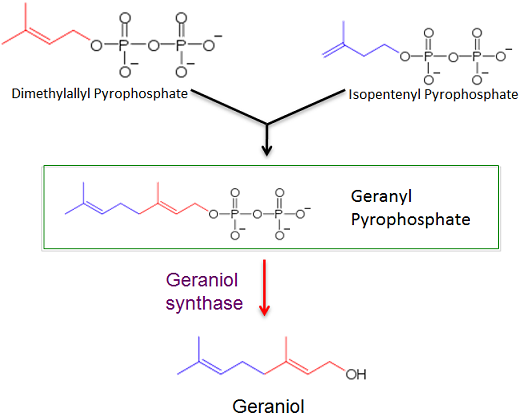
| - | The first purpose for this study is to determine if a gene for geraniol synthase can be properly expressed in the archaea ''Methanococcus maripaludis'' via a plasmid shuttle vector. Successful transformation will result in ''Methanococcus maripaludis'' producing geraniol from geranyl pyrophosphate, an intermediate in a | + | The first purpose for this study is to determine if a gene for geraniol synthase can be properly expressed in the archaea ''Methanococcus maripaludis'' via a plasmid shuttle vector. Successful transformation will result in ''Methanococcus maripaludis'' producing geraniol from geranyl pyrophosphate, an intermediate in a lipid biosynthesis pathway. |
The second plan is to develop novel gene expression and quantification tools for ''Methanogens''. To achieve this we will design novel vectors with the potential to express red fluorescence. This vector will have the additional capability of allowing users to directly fuse their protein of interest with the red fluorescent protein. | The second plan is to develop novel gene expression and quantification tools for ''Methanogens''. To achieve this we will design novel vectors with the potential to express red fluorescence. This vector will have the additional capability of allowing users to directly fuse their protein of interest with the red fluorescent protein. | ||
Revision as of 22:17, 25 September 2013
General Purpose and Goal
The first purpose for this study is to determine if a gene for geraniol synthase can be properly expressed in the archaea Methanococcus maripaludis via a plasmid shuttle vector. Successful transformation will result in Methanococcus maripaludis producing geraniol from geranyl pyrophosphate, an intermediate in a lipid biosynthesis pathway.
The second plan is to develop novel gene expression and quantification tools for Methanogens. To achieve this we will design novel vectors with the potential to express red fluorescence. This vector will have the additional capability of allowing users to directly fuse their protein of interest with the red fluorescent protein.
Objectives
1. This research will create a Methanococcus maripaludis strain that will produce a rose-like fragrance. Salt marshes tend to have a very distinct and bad smell from the production of methane from Methanogens. If Methanococcus maripaludis can successfully produce geraniol then these marshes can be made to have a nice floral fragrance. Because studies have shown that geraniol have inhibited pancreatic and colon cancer growth, this research could be used to produce geraniol for pharmaceutical purposes. The ideal living condition for M. maripaludis is in moderate temperatures, usually around 20-45°C. It has an irregular cocci shape and utilizes flagella for movement. M. maripaludis is an ideal research candidate because of its rapid, reliable growth rate, effective genetic tools and complete genome sequence. (Whitman et al., 1997) The production of geraniol is made possible by the successful expression of a gene coding for geraniol synthase. Geraniol synthase produces geraniol from geraniol diphosphate which is an intermediate in a lipid biosynthesis pathway. To express the geranyl synthase gene a plasmid shuttle vector was created.
2. Our second objective was creating a tool that will allow future researchers and iGEM teams to quantify and regulate gene expression in Archaea.
3. To increase the awareness of synthetic biology among local high school students.
Plan
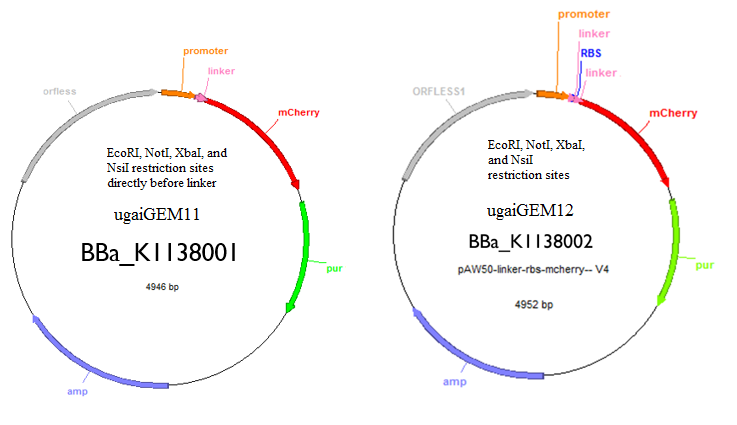
Genetic transformation will be accomplished through the use of a plasmid shuttle vector, pAW50. The DNA sequences encoding for geraniol synthase and mcherry will be added to the pAW50 vector and transformed into E. coli. The sequences were slightly altered for better expression in M. maripaludis by way of codon optimization. This technique involves using the most frequent base pair triplets which appear most often for a certain codon in the M. maripaludis genome. After the genes are inserted into the vector they are transformed into cells of the E. coli strain XL-1 Blue to replicate the plasmid. The E. coli cells will then be lysed to extract the plasmids for transformation into M. maripaludis. The plasmid will also contain a gene encoding for a resistance to the antibiotic puromycin. This will serve as a selection marker in order to confirm successful transformation of the plasmid. Cells which have not taken up a plasmid will die when grown on media that contains puromycin. Fluorescent studies will be accomplished using the plate reader available in Dr. Yan's Lab.
 "
"