Template:Kyoto/Notebook/Aug 27
From 2013.igem.org
(Difference between revisions)
(→Ligation) |
(→Electrophoresis) |
||
| Line 134: | Line 134: | ||
|||100bp ladder||--||-- | |||100bp ladder||--||-- | ||
|- | |- | ||
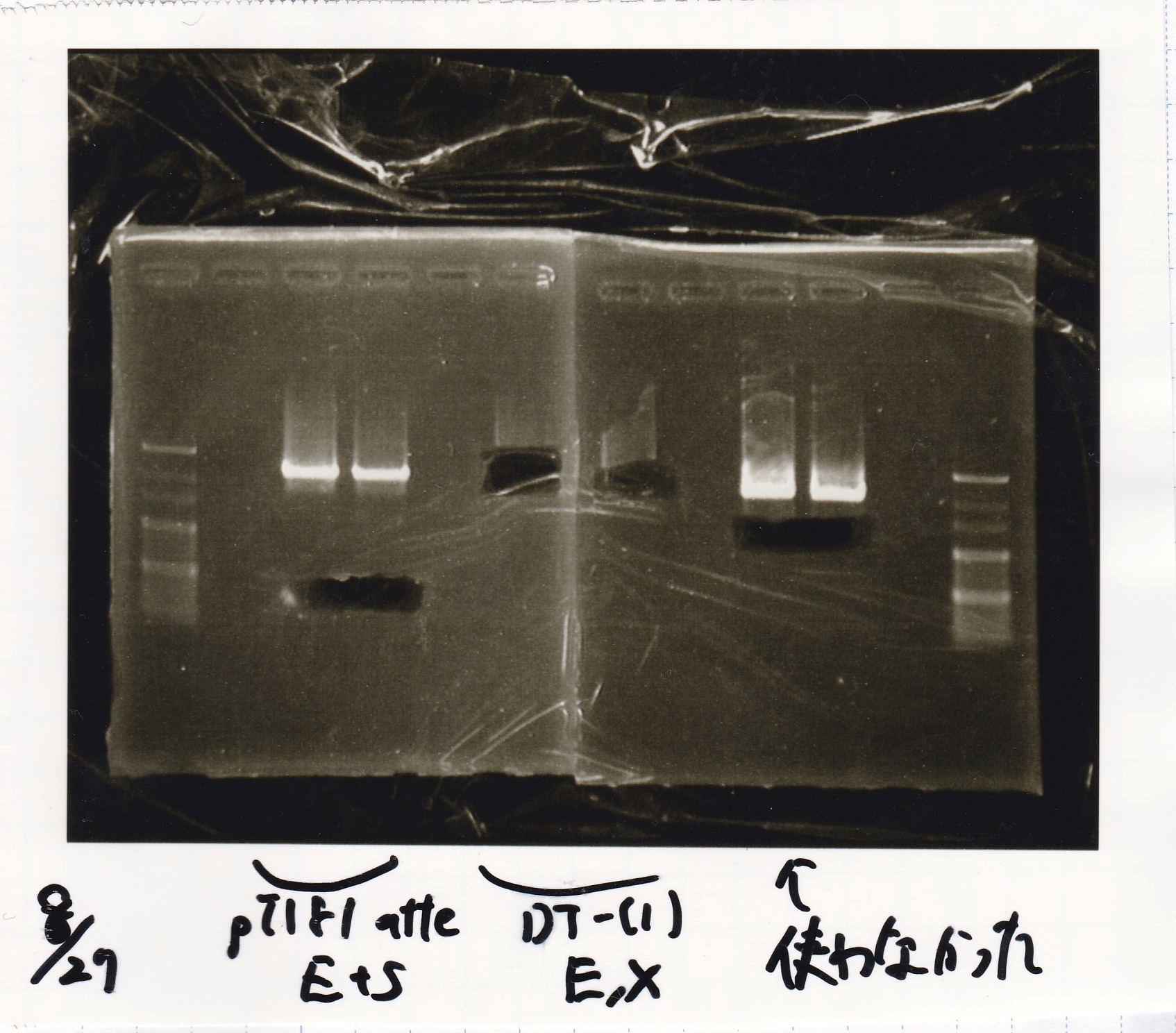
| - | |1||pT181 attenuator|| | + | |1||pT181 attenuator||EcoRI||SpeI |
|- | |- | ||
|2||pT181 attenuator||--||-- | |2||pT181 attenuator||--||-- | ||
Latest revision as of 00:47, 27 September 2013
Contents |
Aug 27
Restriction Enzyme Digestion
| 8/21 pT181 attenuator-1(330µg/mL) | EcoRI | SpeI | 10x BufferB | 100x BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 3.0µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.7µL | 30µL |
| NC | 0.6µL | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL |
| 8/17 DT-1(188µg/mL) | EcoRI | XbaI | 10x BufferB | 100x BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 3.0µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.7µL | 30µL |
| NC | 1.0µl | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL |
| 8/20 Pcon-RBS-luxR-DT-2(344µg/mL) | EcoRI | SpeI | 10x BufferB | 100x BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 2.9µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.8µL | 30µL |
| NC | 0.6µl | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL |
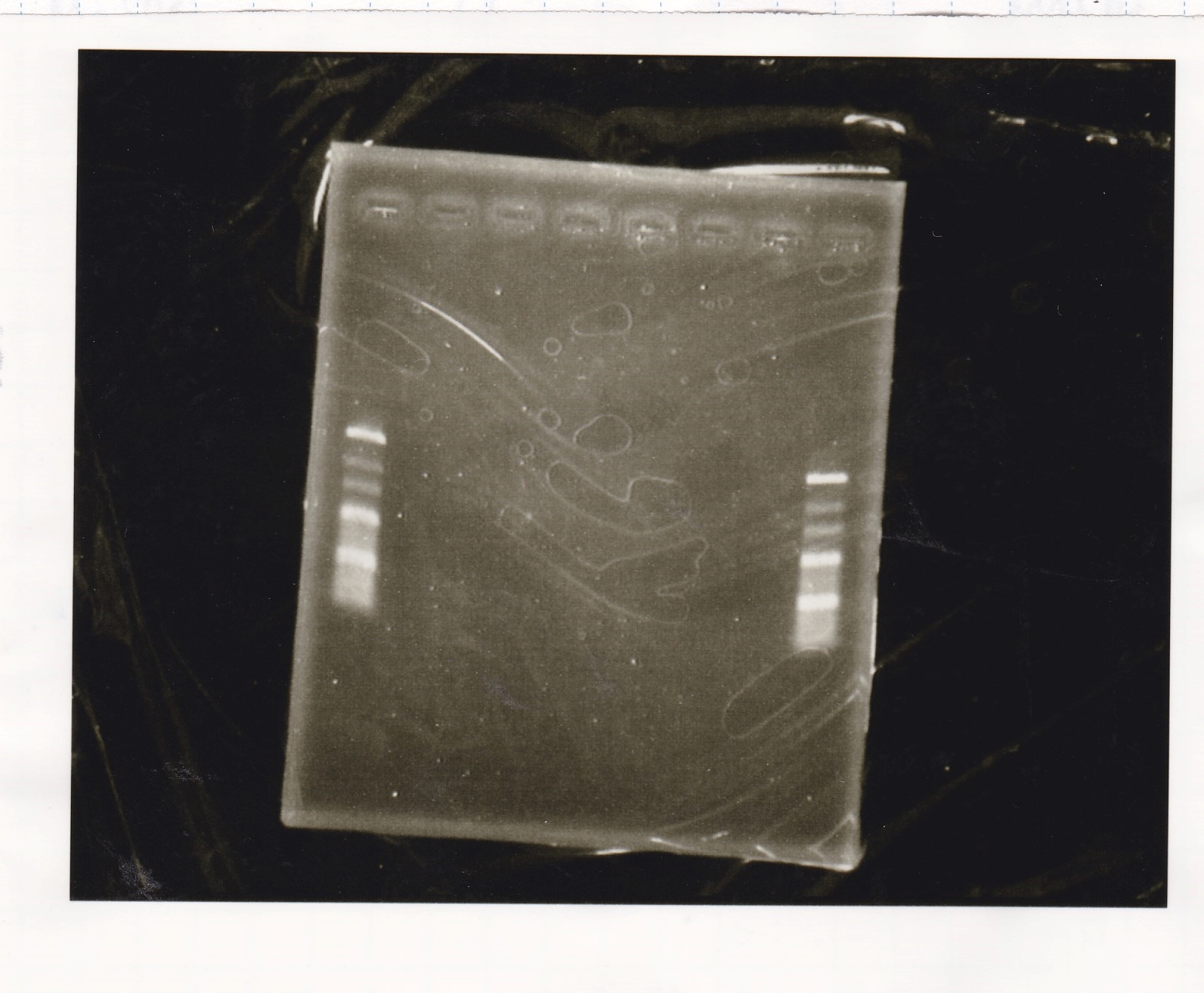
Electrophoresis
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | 100bp ladder | -- | -- |
| 2 | 8/21 pT181 attenuator | EcoI | SpeI |
| 3 | 8/21 pT181 atteniator | -- | -- |
| 4 | 8/17 DT-1 | EcoRI | XbaI |
| 5 | 8/17 DT-1 | -- | -- |
| 6 | 8/20 Pcon-RBS-luxR-DT-2 | EcoI | SpeI |
| 7 | 8/20 Pcon-RBS-luxR-DT-2 | -- | -- |
| 8 | 100bp ladder | -- | -- |
Colony PCR
Liquid Culture
| Sample | medium |
|---|---|
| 8/26 Pλ-RBS-luxI-DT-1 | Plusgrow medium (+Amp) |
| 8/26 Pλ-RBS-luxI-DT-2 | Plusgrow medium (+Amp) |
Restriction Enzyme Digestion
| 8/21 pT181 attenuator-1(330µg/mL) | EcoRI | SpeI | 10x BufferB | 100x BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 3.0µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.7µL | 30µL |
| NC | 0.6µL | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL |
| 8/17 DT-1(188µg/mL) | EcoRI | XbaI | 10x BufferB | 100x BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 3.0µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.7µL | 30µL |
| NC | 1.0µl | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL |
| 8/20 Pcon-RBS-luxR-DT-2(344µg/mL) | EcoRI | SpeI | 10x BufferB | 100x BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 2.9µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.8µL | 30µL |
| NC | 0.6µl | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL |
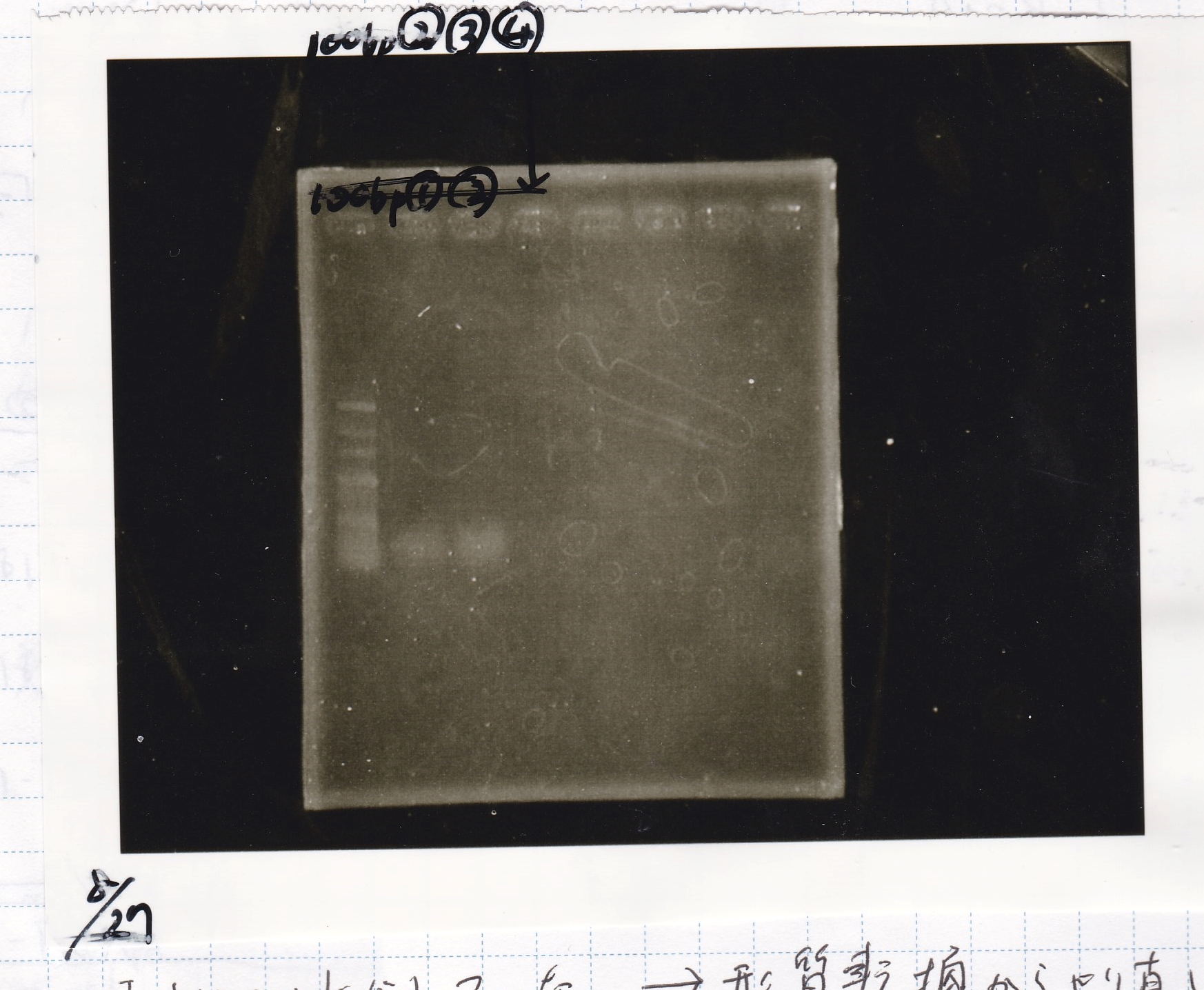
Electrophoresis
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 100bp ladder | -- | -- | |
| 1 | pT181 attenuator | EcoRI | SpeI |
| 2 | pT181 attenuator | -- | -- |
| 3 | DT-(1) | EcoRI | XbaI |
| 4 | DT-(1) | -- | -- |
| 5 | Pcon-RBS-luxR-DT-(2) | EcoRI | SpeI |
| 6 | Pcon-RBS-luxR-DT-(2) | -- | -- |
| 100bp ladder | -- | -- |
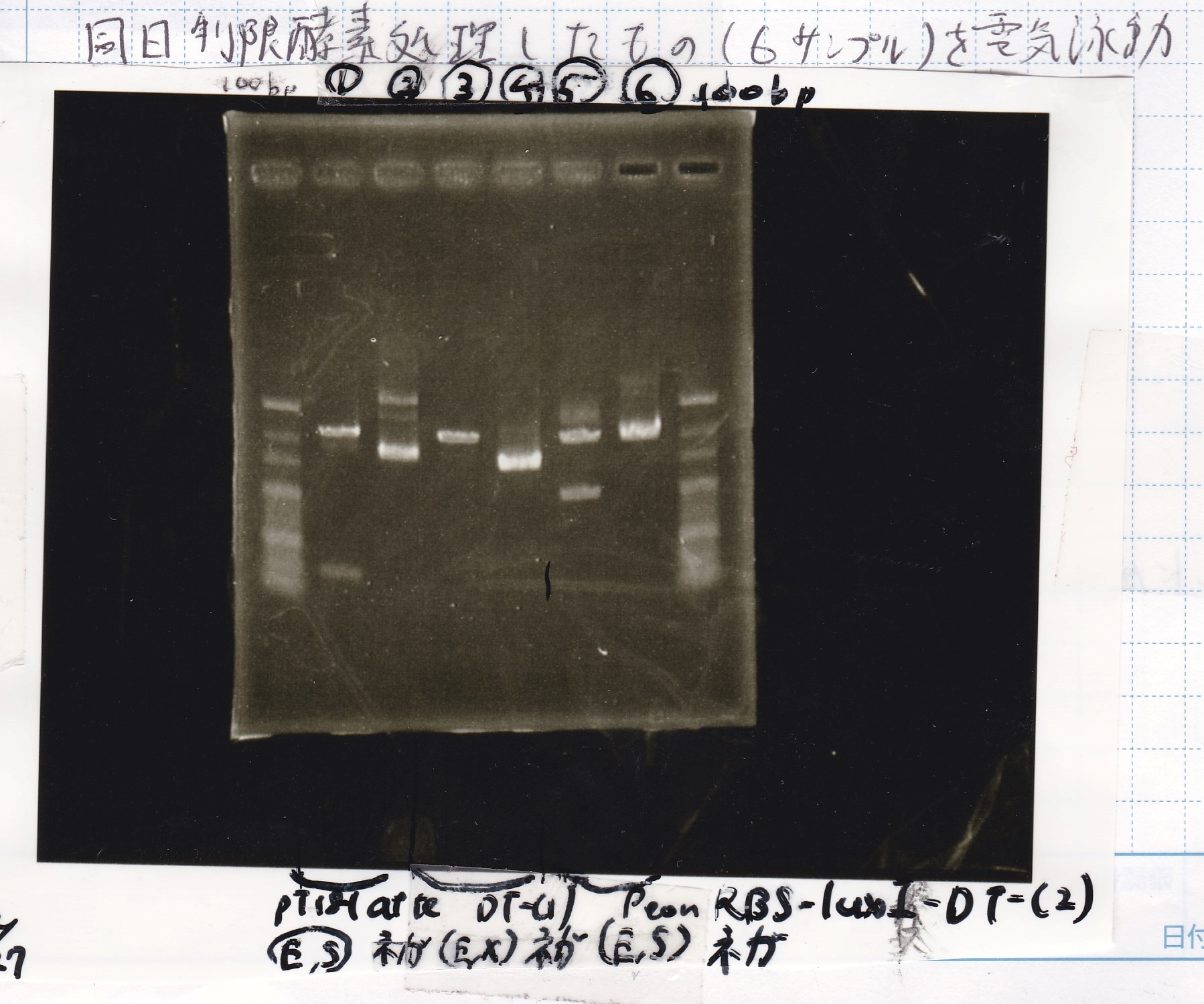
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 100bpladder | -- |
| 2 | pT181 attenuator-1 | EcoRI&SpeI |
| 3 | pT181 attenuator-1 | EcoRI&SpeI |
| 4 | DT-1 | EcoRI&XbaI |
| 5 | DT-1 | EcoRI&XbaI |
| 6 | Pcon-RBS-luxR-DT-2 | EcoRI&SpeI |
| 7 | Pcon-RBS-luxR-DT-2 | EcoRI&SpeI |
| 8 | 100bpladder | -- |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| pT181 attenuator-1(EcoRI&SpeI) | 4.2 | 1.62 | 0.29 |
| DT-1(EcoRI&XbaI) | 17 | 1.88 | 0.88 |
Transformation
| Name | Sample | Competent Cells | Total | Plate |
|---|---|---|---|---|
| 8/26 tetR aptamer 12_1R-DT | 1µL | 10µL | 11µL | CP |
| 8/26 pT181 attenuator-DT | 1µL | 10µL | 11µL | CP |
| 8/26 pT181 antisense-DT | 1µL | 10µL | 11µL | CP |
| 8/26 Spinach-DT | 1µL | 10µL | 11µL | CP |
| 8/26 Plac-pT181 attenuator | 1µL | 10µL | 11µL | CP |
| 8/26 Pcon-pT181 attenuator | 1µL | 10µL | 11µL | Amp |
| 8/26 Plac-pT181 antisense | 1µL | 10µL | 11µL | CP |
| 8/26 Pcon-pT181 antisense | 1µL | 10µL | 11µL | Amp |
Liquid culture
| Sample | medium |
|---|---|
| 8/16 Plux | Plusgrow medium(+CP) |
Genome PCR
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | SasA_fwd primer | SasA_rev primer | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16 | 25 |
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | RpaA_fwd primer | RpaA_rev primer | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16 | 25 |
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | RpaB_fwd primer | RpaB_rev primer | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16 | 25 |
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | PkaiBC_fwd primer | PkaiBC_rev primer | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 50°C | 68°C | -- |
| 2 min | 10 sec | 30 sec | 38 sec | 30 cycles |
Ligation
| state | Vector | Inserter | ||
|---|---|---|---|---|
| experiment | 8/27 DT (EcoRI & XbaI) | 2.8 | 8/21 RBS-lysis3 (EcoRI & SpeI) | |
| experiment | 8/27 DT (EcoRI & XbaI) | 2.8 | 8/27 pT181 attenuator (EcoRI & SpeI) | |
- Samples were evaporeted used evaporator into about 3 µL.
| sample | MilliQ | Ligation High | total |
|---|---|---|---|
| 3 | 4 | 3.5 | 10.5 |
- incubate overnight at 4 °C
 "
"