Team:Stanford-Brown/Projects/EuCROPIS
From 2013.igem.org
(→Data) |
(→Data) |
||
| Line 22: | Line 22: | ||
== '''Data''' == | == '''Data''' == | ||
| - | <gallery | + | <gallery mode=packed-hover widths="180px" heights="120px" perrow="3"> |
Image: 0012.jpg|'''0012 - clump of WT bacillus (phase contrast)<br>''' | Image: 0012.jpg|'''0012 - clump of WT bacillus (phase contrast)<br>''' | ||
Image:RyanGallagher.jpg|'''0013 - Spores + Vegetative bacillus<br>''' | Image:RyanGallagher.jpg|'''0013 - Spores + Vegetative bacillus<br>''' | ||
Revision as of 06:59, 27 September 2013
Contents |
Introduction
Imagine taking synthetic biology to space. In 2011, the Brown-Stanford iGEM team did just that, designing a synthetic-biology based Mars colony. At the center of the concept was PowerCell, a consortium of a photosynthetic, diazotrophic (nitrogen-fixing) cyanobacterium engineered to secrete sucrose, and a "production cell".
Construct Assembly
We began by researching and identifying genes associated with sporulation, germination, and sucrose induction in Bacillus subtilis. We decided to focus on sacY, a gene the regulates sucrose induction and is itself induced by the presence of sucrose, and spo0A, a gene that regulates sporulation by influencing more than 500 genes related to sporulation. We attempted to isolate the genes from the genome using PCR with mixed results. We decided instead to construct the two promoters by annealing complimentary oligos from Elim, and using PCR to add the BioBrick ends.
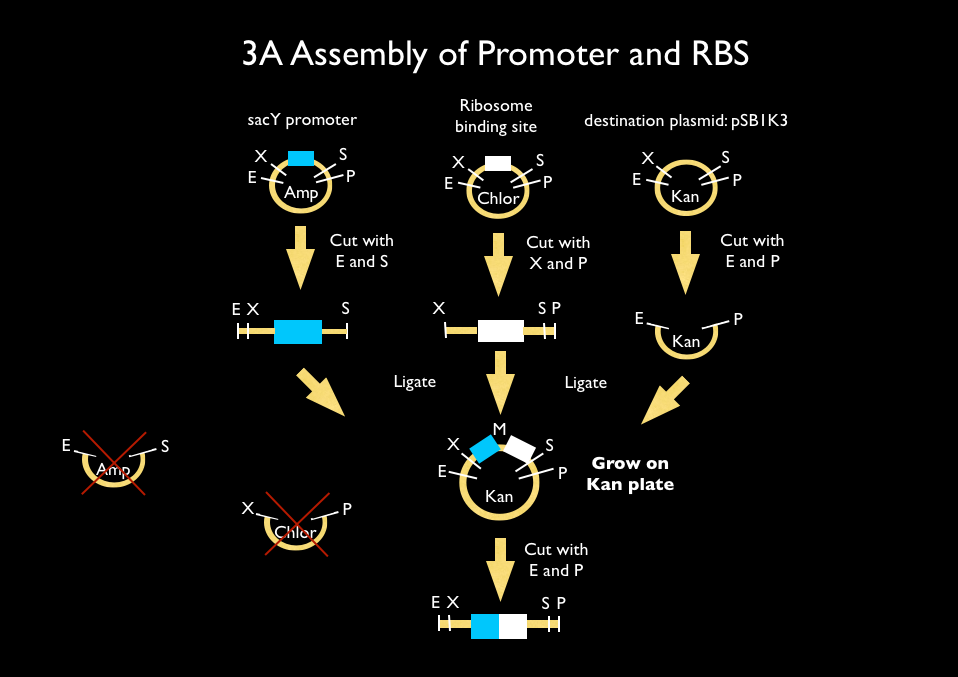
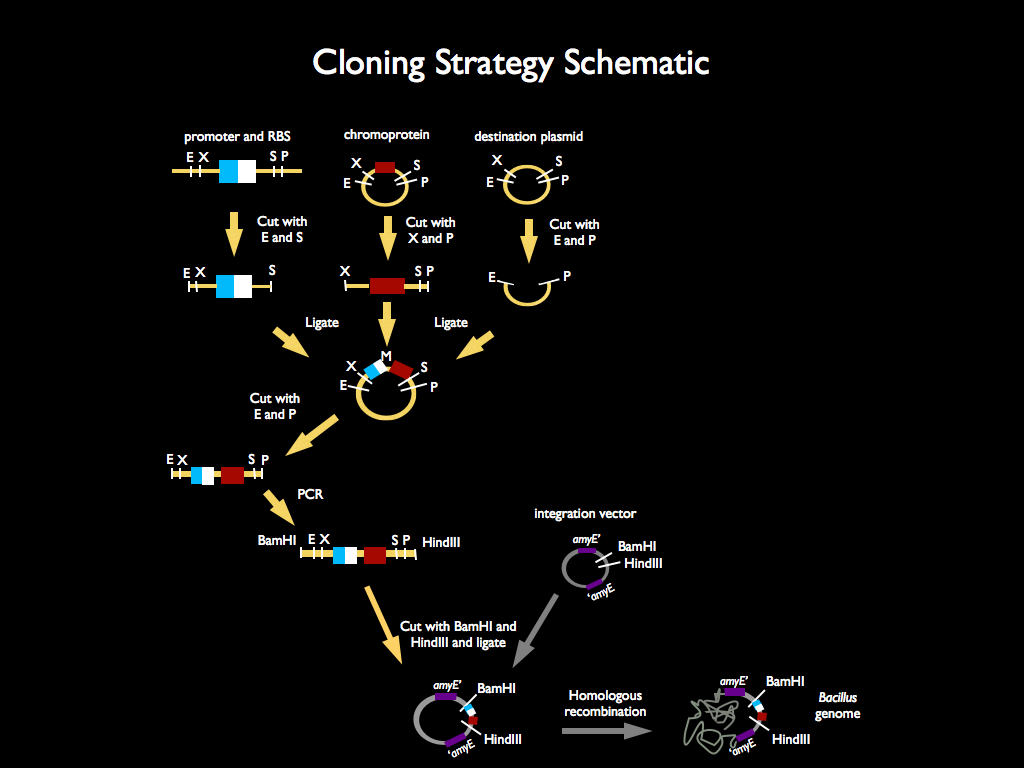
We used 3A assembly to add a ribosome binding site from the registry (BBa_B0034) directly downstream of our sucrose inducer promoter (see Figure A). After an E/P digestion, we proceeded to another 3A assembly to add a red chromoprotein (eForRed: BBa_K592012) downstream of the sucrose inducer promoter and ribosome binding site (Figure B). Likewise we did another assembly in parallel to add a red fluorescent protein (BBa_E1010) downstream of the sucrose inducer promoter and ribosome binding site. We then used primers from Elim and PCR to add the restriction sites BamHI and HindIII to either side of our two constructs. In addition, we ordered two constructs from DNA 2.0: the sucrose inducing promoter linked to eForRed and the sporulation regulator promoter linked to a blue chromoprotein (BBa_K864401). We designed the constructs such that they were flanked with the restriction sites BamHI and HindIII. These two restriction sites allowed us to ligate each of our constructs into Bacillus subtilis integration vectors (BGSC: ECE115). Finally, we used electroporation to transform into electrocompetent Bacillus subtilis cells.
Lab Notebook
Our team diligently kept a lab notebook throughout the summer. The full notebook can be accessed here.
Data
BioBricks
[http://parts.igem.org/Part:BBa_K1218001 BBa_K1218001 (pSB1C3)]: This part is the sacY promoter; sacY is a gene that regulates sucrose induction in Bacillus subtilis.
[http://parts.igem.org/Part:BBa_K1218020 BBa_K1218020 (pSB1K3)]: This is a compound part containing the sacY promoter (BBa_K1218001: a sucrose inducer in Bacillus subtilis) and a ribosome binding site (BBa_B0034).
[http://parts.igem.org/Part:BBa_K1218021 BBa_K1218021 (pSB1C3)]: This part is the spo0A promoter; spo0A regulates sporulation in Bacillus subtilis.
[http://parts.igem.org/Part:BBa_K1218023 BBa_K1218023 (pJ201)]: This composite part contains sacY (BBa_K1218001: a sucrose inducer in Bacillus subtilis), a ribosome binding site (BBa_B0034), and eForRed, a red chromoprotein developed by Uppsala ([http://parts.igem.org/Part:BBa_K592012 BBa_K592012]). We intend to use it as a chromogenic reporter induced by sucrose.
[http://parts.igem.org/Part:BBa_K1218025 BBa_K1218025 (pSB1K3)]: This composite part contains sacY (BBa_K1218001: a sucrose inducer in Bacillus subtilis), a ribosome binding site (BBa_B0034), and a red fluorescent protein (BBa_E1010). We intend to use it as a chromogenic reporter induced by sucrose.
Acknowledgements
We would like to thank the many people who made this project possible.
- Ryan Kent, Dr. Lynn Rothschild, Dr. Joe Shih, and Dr. Kosuke Fujisima -- Primary advisors
- Dr. Gary Wessel -- Advice on testing construct
- Dr. Rocco Mancinelli -- Feedback on our presentation, papers about the EuCROPIS mission
- Dr. Lilah Rahn-Lee -- Advice on choosing B. subtilis genes
- Dr. Daniel R. Zeigler, BGSC Director and the Bacillus Genetic Stock Center -- Free strains of Bacillus integration vectors
- Dr. Elwood Agasid -- Explanation of EuCROPIS mission from an engineering perspective
 "
"