From 2013.igem.org
(Difference between revisions)
|
|
| Line 294: |
Line 294: |
| | </div> | | </div> |
| | | | |
| - | ===PCR=== | + | ===Sequence PCR=== |
| | <div class="experiment"> | | <div class="experiment"> |
| | <span class="author">Tatsui</span> | | <span class="author">Tatsui</span> |
Revision as of 17:24, 27 September 2013
Sep 14
Restriction Enzyme Digestion
Hirano
| | 9/14 pSB4K5S | EcoRI | XbaI | SpeI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2cuts | 10µL | 1µL | 0µL | 1µL | 0µL | 3µL | 0µL | 3µL | 12µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 1µL | 7.5µL | 10µL
|
| | 9/13 Plac | XbaI | PstI | BufferH | BSA | MilliQ | total
|
| 1 cut | 9µL | 0µL | 1µL | 3µL | 3µL | 14µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 1µL | 1µL | 7µL | 10µL
|
| | 9/14 PKaiBC | EcoRI | XbaI | SpeI | PstI | BufferB | BSA | MilliQ | total
|
| 2cuts | 11µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 11µL | 30µL
|
| NC | 4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 4µL | 10µL
|
| | 9/14 RpaB | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total
|
| 2cuts | 13µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 9µL | 30µL
|
| NC | 2µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 5µL | 10µL
|
| | 8/17 RBS-GFP-DT | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total
|
| 1cut | 9µL | 0µL | 1µL | 0µL | 0µL | 3µL | 3µL | 14µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
Liquid Culture
Hirano
| Sample | medium
|
| 9/11 Plac(BBa-R0011)-(3) | Plusgrow medium(+Amp)
|
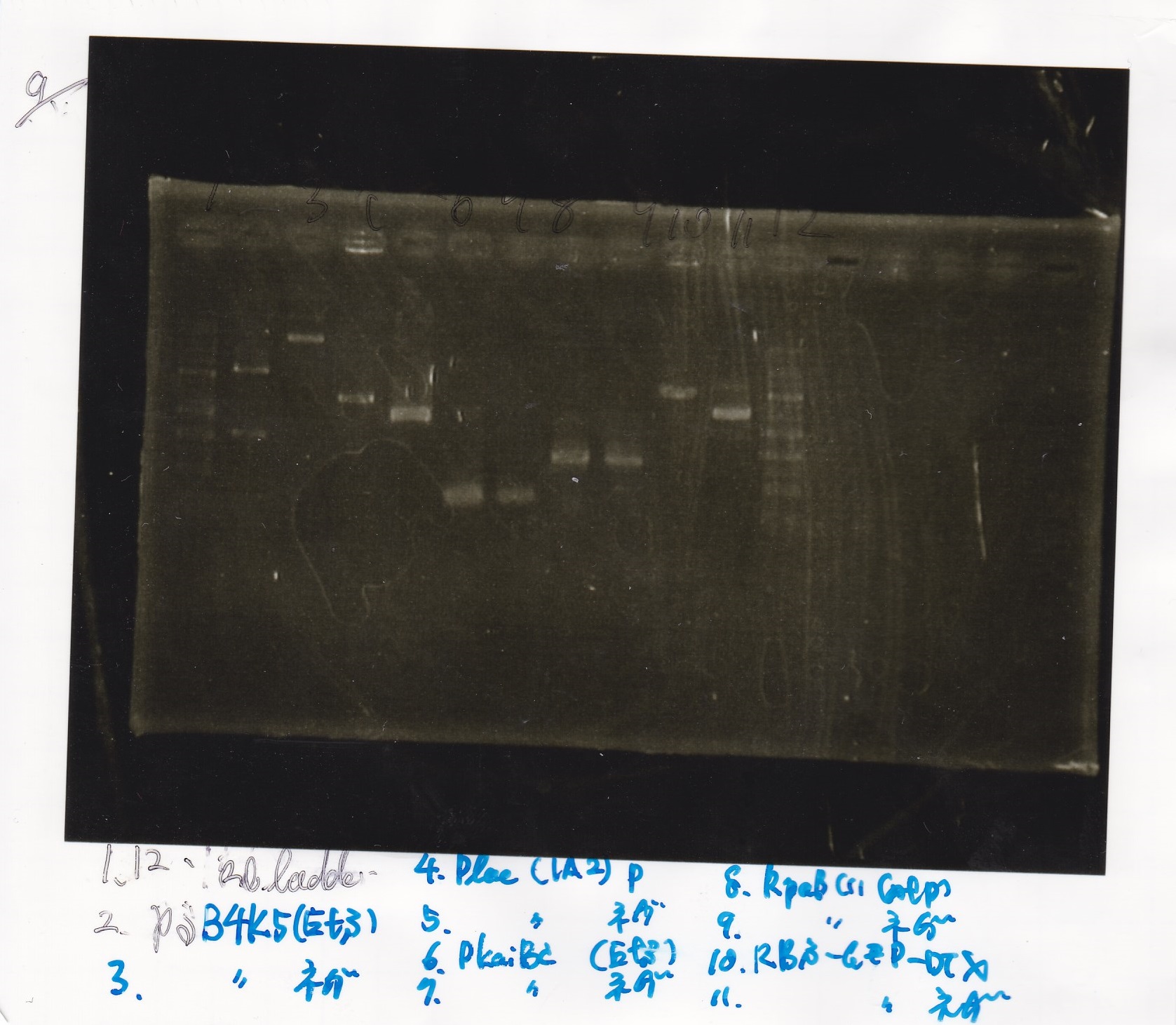
Electrophoresis
Hirano
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kb ladder | -- | --
|
| 2 | pSB4K5(1) | EcoRI | SpeI
|
| 3 | pSB4K5(1) | -- | --
|
| 4 | Plac(1A2) | PstI | --
|
| 5 | Plac(1A2) | -- | --
|
| 6 | PKaiBC | EcoRI | SpeI
|
| 7 | PKaiBC | -- | --
|
| 8 | RpaB(1) | XbaI | PstI
|
| 9 | RpaB(1) | -- | --
|
| 10 | RBS-GFP-DT | XbaI | --
|
| 11 | RBS-GFP-DT | -- | --
|
| 12 | 1kb ladder | -- | --
|
| Lane | Sample | Enzyme
|
| 1 | 1kb ladder | --
|
| 2 | Plac(pSB1A2) | PstI
|
| 3 | Plac(pSB1A2) | --
|

Sequence PCR
No name
| genome DNA | Big Dye | 5x buffer | temp(400mg) | primer | milliQ
|
| Pcon-RBS-luxR-DT | 0.5 | 1.75 | 0.9 | 0.5 | 6.85
|
| Pcon-RBS-luxR-DT | 0.5 | 1.75 | 0.4 | 0.5 | 7.35
|
| Pcon-RBS-tetR-DT | 0.5 | 1.75 | 3 | 0.5 | 5.25
|
| Pcon-RBS-tetR-DT | 0.5 | 1.75 | 3 | 0.5 | 5.25
|
| Plux | 0.5 | 1.75 | 2.4 | 0.5 | 5.35
|
| Plux-RBS-GFP-DT | 0.5 | 1.75 | 1.9 | 0.5 | 5.85
|
| Plux-RBS-GFP-DT | 0.5 | 0.5 | 1.9 | 0.5 | 5.85
|
| Pcon-RBS-GFP-DT | 0.5 | 0.5 | 1.2 | 0.5 | 6.55
|
| Pcon-RBS-GFP-DT | 0.5 | 0.5 | 1.2 | 0.5 | 6.55
|
| RBS-lysis3-DT | 0.5 | 0.5 | 1.0 | 0.5 | 6.75
|
| Pcon-antisense-Spinach-DT | 0.5 | 0.5 | 1.4 | 0.5 | 6.35
|
| Pcon-RBS lacZα-DT | 0.5 | 0.5 | 1.4 | 0.5 | 6.35
|
| Ptet-RBS lacZα-DT | 0.5 | 0.5 | 0.59 | 0.5 | 1.85
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 96°C | 96°C | 50°C | 60°C | --
|
| 2min | 10sec | 5sec | 4min | 30cycle
|
Restriction Enzyme Digestion
No name
| | 9/14 Plac(1A2) | EcoRI | XbaI | SpeI | PstI | BufferB | BSA | MilliQ | total
|
| 2cuts | 23µL | 0µL | 0µL | 0.3µL | 0µL | 3µL | 3µL | 0µL | 30µL
|
| | 9/14 RBS-GFP-DT | EcoRI | XbaI | SpeI | PstI | BufferH | BSA | MilliQ | total
|
| 2 cuts | 23µL | 1µL | 0µL | 0µL | 0µL | 3µL | 3µL | 0µL | 30µL
|
Ligation
Nakamoto and Hirano
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/13 Pcon-PT181attenuator(SpeI&PstI) | 1.7 | 9/13 RBS-lacZα-DT (XbaI & PstI) | 12.5 | 5
|
| experiment | 9/13 pSB2C3 (XbaI & PstI) | 1.9 | 9/14 RpaB (XbaI & PstI) | 16.3 | 10.5
|
| experiment | 9/13 pSB2C3 (XbaI & PstI) | 4.7 | 9/14 PkaiBC (EcoRI & SpeI) | 6.7 | 5.7
|
| experiment | 9/14 RBS-GFP-DT (PstI&XbaI) | 0.7 | 9/14 PkaiBC (EcoRI & SpeI) | 2.6 | 1.6
|
| experiment | 9/14 Plac (SpeI&PstI) | 2.4 | 9/14 RBS-lysis1-DT (XbaI & PstI) | 4.9 | 3.6
|
| experiment | 9/14 Plac (SpeI&PstI) | 2.4 | 9/14 RBS-lysis2-DT (XbaI & PstI) | 7.0 | 4.7
|
| experiment | 9/14 Plac (SpeI&PstI) | 2.4 | 9/14 RBS-lysis3-DT (XbaI & PstI) | 9.2 | 5.8
|
Samples were evaporeted used evaporator into about 7 µL
Colony PCR
No name
| Sample | base pair
|
| 9/13 Plux+RBS-lysis1-DT (1)~(3) | --
|
| 9/13 Plux+RBS-lysis2-DT (1)~(3) | -
|
| 9/13 Plux+RBS-lysis3-DT (1)~(2) | -
|
| Sample | base pair
|
| 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT (1)~(4) | --
|
| 9/13 Pcon-PT181attenuator+aptamer121R-DT (1)~(2) | -
|
| 9/13 Plac+PT181attenuator (1) | -
|
| Sample | base pair
|
| Dλ-lux(1) (1)~(2) | --
|
| NC | --
|
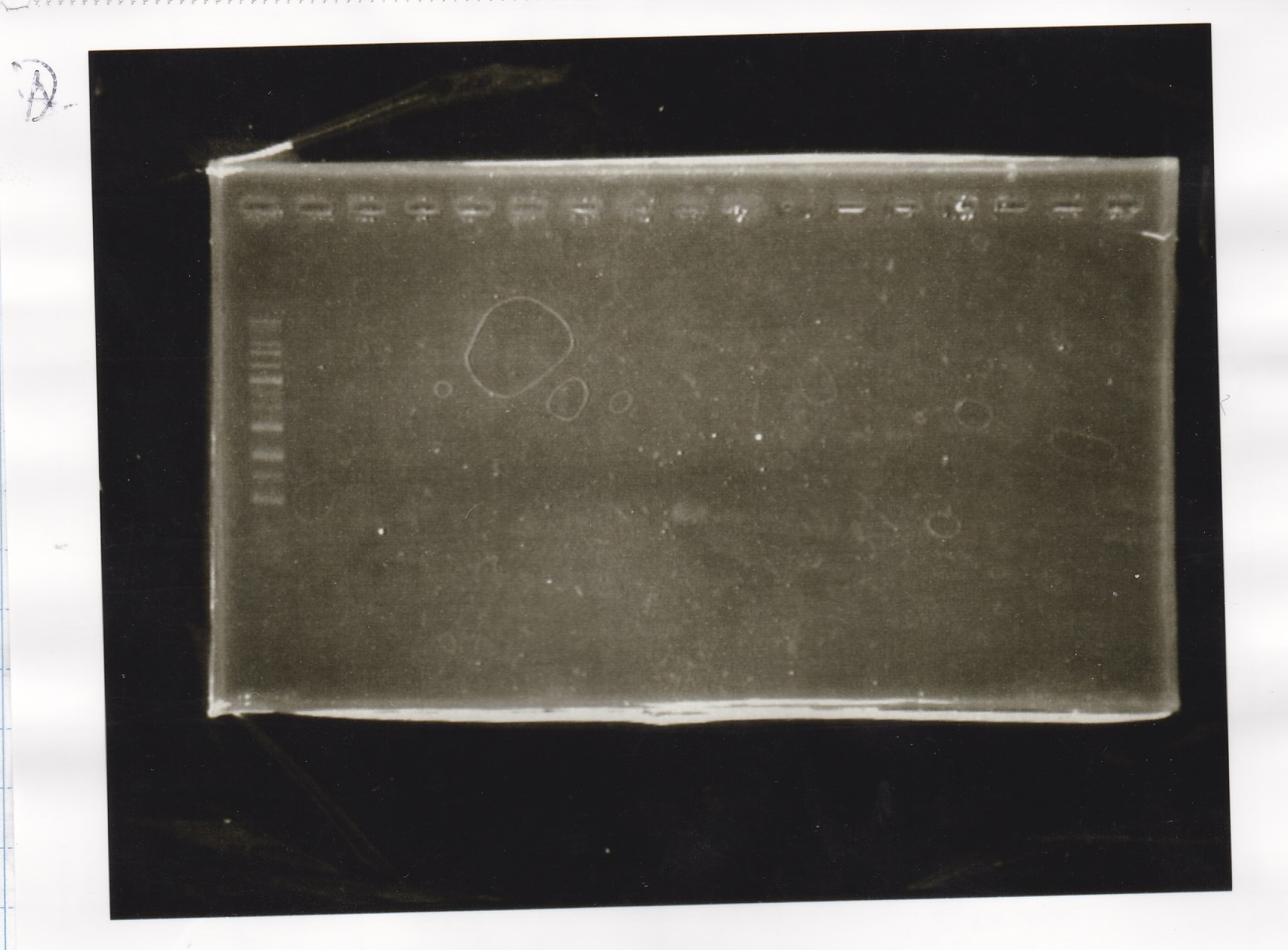
Electrophoresis
No name
| Lane | Sample
|
| 1 | 1kbp ladder
|
| 2 | 9/13 Plux+RBS-lysis1-DT1
|
| 3 | 9/13 Plux+RBS-lysis1-DT2
|
| 4 | 9/13 Plux+RBS-lysis1-DT3
|
| 5 | 9/13 Plux+RBS-lysis2-DT1
|
| 6 | 9/13 Plux+RBS-lysis2-DT2
|
| 7 | 9/13 Plux+RBS-lysis2-DT3
|
| 8 | 9/13 Plux+RBS-lysis3-DT1
|
| 9 | 9/13 Plux+RBS-lysis3-DT2
|
| 10 | 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT1
|
| 11 | 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT2
|
| 12 | 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT3
|
| 13 | 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT4
|
| 14 | 9/13 Ptet-pT181 antisense+Spinach-DT1
|
| 15 | 9/13 Pcon-pT181 attenuator+aptamer12-1R-PT1
|
| 16 | 9/13 Pcon-pT181 attenuator+aptamer12-1R-PT1
|
| 17 | 9/13 Plac+pT181 attenuator
|
| Lane | Sample
|
| 1 | 1kbp ladder
|
| 2 | Pλ-lux(1)1
|
| 3 | Pλ-lux(1)1
|
| 4 | NC
|

Liquid Culture
No name
| Sample | medium
|
| 9/13 Pλ-luxI(1)-1 | Plusgrow medium(+Amp)
|
Transformation
Nakamoto
| Name | Sample | Competent Cells | Total | Plate
|
| 9/13 RBS-lysisZα-DT(XbaI&PstI)+9/13 Pcon-PT181 attenuator(SpeI&PstI) | 2µL | 20µL | 22µL | Amp
|
| 9/13 SB2C3+9/14 PzαB | 2µL | 20µL | 22µL | CP
|
| 9/13 pSB2C3+9/14 PkaiBC | 2µL | 20µL | 22µL | CP
|
| 9/13 RBS-GFP-DT+9/14 PkaiBC | 2µL | 20µL | 22µL | CP
|
| Plac+RBS-lysis1-DT | 2µL | 20µL | 22µL | Amp
|
| Plac+RBS-lysis2-DT | 2µL | 20µL | 22µL | Amp
|
| Plac+RBS-lysis2-DT | 2µL | 20µL | 22µL | Amp
|
Sequence PCR
Tatsui
| genome DNA | temp | Big Dye | 5x buffer | primer | MilliQ
|
| 9/6 PT181 attenuator(VR) | 3.9 | 0.5 | 1.75 | 0.5 | 3.85
|
| 8/17 RBS lacZα-DT | 2.1 | 0.5 | 1.75 | 0.5 | 5.65
|
| 9/6 PT181 attenuator(VR) | 3.9 | 0.5 | 1.75 | 0.5 | 3.85
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 96°C | 50°C | 60°C | 4°C | --
|
| 10sec | 5sec | 4min | 1min | 30cycle
|
 "
"



