Team:Biwako Nagahama/Project
From 2013.igem.org
(→Binary Vector) |
(→E.coli-ink) |
||
| Line 173: | Line 173: | ||
<p>Lane1:JM109 with pUC18 Lane2:JM109 with pUC18/Mb</p> | <p>Lane1:JM109 with pUC18 Lane2:JM109 with pUC18/Mb</p> | ||
<p>Next, We have to mutate this DNA fragment encoding myoglonbin because that includes EcoRⅠand XbaⅠsite. GAATTC that is EcoRⅠsite is changed to GAATTT. TTC is encoding amino acid and TTT too. As same CATT that is XbaⅠsite is changed to .</p> | <p>Next, We have to mutate this DNA fragment encoding myoglonbin because that includes EcoRⅠand XbaⅠsite. GAATTC that is EcoRⅠsite is changed to GAATTT. TTC is encoding amino acid and TTT too. As same CATT that is XbaⅠsite is changed to .</p> | ||
| - | <p>EcoRⅠsite pointmutation protocol</p> | + | <p>EcoRⅠsite pointmutation protocol below</p> |
| - | <p></p> | + | <p>First,We design the primer occurring pointmutation.Primer sequence below.</p> |
| - | <p></p> | + | <p>F1</p> |
| - | <p></p> | + | <p>R1</p> |
| + | <p>F2</p> | ||
| + | <p>R2</p> | ||
| + | <p>F3</p> | ||
| + | <p>R3</p> | ||
| + | |||
| + | <p>We use primer F1 and R2 to make the left end fragment on PCR. PCR product</p> | ||
| + | |||
| + | <p>We use primer F2 and R3 to make middle fragment on PCR.</p> | ||
| + | <p>We use thermal cycler on PCR. but,middle fragment theoretical value is about 60 base pair, it is very short. So this PCR don’t amplify DNA fragment we targeting but plasmid itself. PCR enzyme using by us is Prime star Max. Prime star Max is highly efficient enzyme. We replace Prime star Max with prime star hot start , but result is same. </p> | ||
| + | <p>Hereupon we hit upon an idea. We worked PCR by hand with water bath at 55℃, 70℃ and 98℃ because We think thermal cycler appear gradient temperature and this is a cause of failure.. We dip sample in water bath one by one. 98℃,55℃,70℃,98℃,55℃・・・・.</p> | ||
<p></p> | <p></p> | ||
<p></p> | <p></p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
Revision as of 20:03, 27 September 2013
Contents |
AgRePaper&E.coli-ink
Cellulose is used as raw material for paper, so our team experimented various ways to increase the amount of cellulose produced by agrobacterium and using it to make papers. For this we developed the different parts to insert into the system of agrobacterium. Among them are the genes used for expression of the curdlan. Similarly, genetic parts in order to increase the expression of the cellulose, along with the agrobacterium type binary vector were also developed . We are also working on recycling the produced paper by degrading the cellulose to D-Glucose using various enzymes. We worked for the preparation of the biological ink using the sperm whale's cells by genetically modification to increase amount of myoglobin. Then, we observed the change on the color of the product by altering the formation of myoglobin and the production amount of myoglobin with the insertion of T7 promoter to the cell system.
Agrepaper
Background
Today, Our Earth is confronted with global warming and depletion of ozone layer, organic extinction, an energy problem as an environmental issue.
We think it is caused by deforestation. We usually use paper. For example, writing letter and Printing papers newspaper, origami, etc… Many woods have felled for such paper.
To solve that issue, Make paper from except woods and raw affected environment material. So, Many woods don’t have to fell for make paper.
How to solve it?
First, I need to study Paper.
Main ingredient of plant fiber is cellulose. Particularly, Ingredient of Paper is Cellulose and Hemicellulose, Lignin. Cellulose work as scaffold, Hemicellulose work as adhesion, Lignin work as Space-filling.
Cellulose and Hemicellulose is from not woods but synthetic biology. So, the issue probably solved.
Secretion of fibrils from Agro
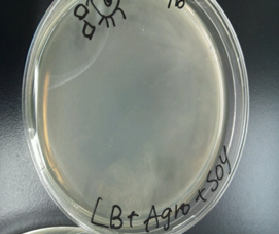
For this year’s project our team aimed at making the paper by using Agro bacterium. So, we confirmed whether carbohydrate developed from Agrobacterium could be used as the raw material for making paper.
Materials
Agro bacterium tumefaciens C58
Agro bacterium tumefaciens C58 bacterial liquid
+LB medium 2 ml
+ampicillin 2 µl
+Agro bacterium tumefaciens C58
Congo red
LB medium
+1.0%(w/v) Triptone 2 g
+0.5%(w/v) yeast extract 1 g
+1.0%(w/v) NaCl 2 g
Experiment1
Sample 1 curdlan 15 µl +Congo red 5 µl +Agro bacterium precipitation
Sample 2 de-ionized water 15 µl +Congo red 5 µl +Agro bacterium precipitation
We centrifuged Sample 1 and 2 in 5 minutes with cooled centrifuges(4 ℃,13000 rpm).
As a result, Sample 1 had red precipitates. Sample 2 rationally melted.
So, we confirmed the presence of carbohydrate in the precipitates obtained from Agro bacterium.
Experiment2
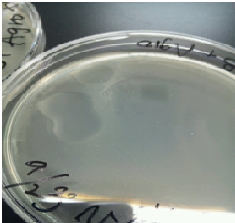
Next, we found that carbohydrate could be produced when soy milk is added to the fluid bacteria
So, we experimented for the confirmation of the above findings.
Sample 3 LB medium 7 ml +Agro bacterium+soy 40 µl
Sample 4 LB medium 7 ml +Agro bacterium
Sample 5 LB medium 7 ml +soy 40 µl
Sample 6 LB medium 7 ml
We cultivated Sample 3 ~ 6 at 28 ℃.for 12 hours
Then, Sample1 and 2 produced a white film like substance but Sample 3 and 4 did not.
We put the white films from Sample 1 and 2 into micro-tubes.
Sample3 Sample4
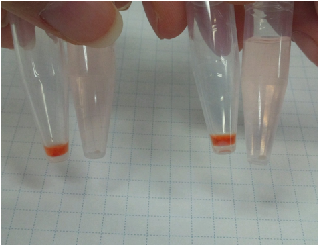
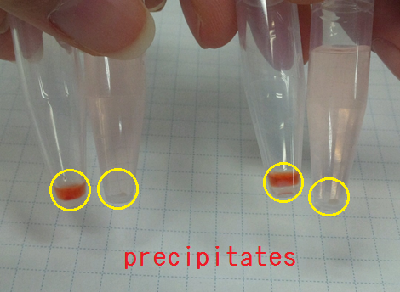
Sample 7 supernatant(Sample 1) 15 µl +Congo red 5 µl
Sample 8 supernatant(Sample 1) 15 µl +Congo red 5 µl +de-ionized water 1 ml
Sample 9 supernatant(Sample 2) 15 µl +Congo red 5 µl
Sample 10 supernatant(Sample 2) 15 µl +Congo red 5 µl +de-ionized water 1 ml
Sample 11 Congo red 5 µl +de-ionized water 1 ml
We centrifuged all the samples for 5 minutes at 4 ℃,13000 rpm.
As a result, Sample 7 ~ 10 gave red precipitates but Sample 11 did not.
So, we confirmed Sample 7 ~ 10 had carbohydrate.
Through these experiments, we confirmed that Agro bacterium can discharge carbohydrate without soy milk.
Reference
1, Laboratory maintenance of Agrobacterium.
(http://europepmc.org/articles/PMC3350319)
2, Coordination of Division and Development Influences Complex Multicellular Behavior in Agrobacterium tumefaciens (Jinwoo Kim.¤, Jason E. Heindl., Clay Fuqua)
3, Kirin Kyowa Foods Company (http://www.kirinkyowa-foods.co.jp/products/curdlan/)
4, Howard Hughes Molecular Biology Summer Research Program Poster, Austin, TX , August, 1995 Microcopy of Curdlan Structure
5, Iain M. Cheeseman and R.Malcom Brown,Jr, Department of Botanty , The University of Texas at Austin,TX,78713
(http://www.botanty.utexas.edu/facstaff/facpages/mbroun/ongres/icheeze.htm)
About curdlan
Agrobacterim makes carbohydrate chain. For exampe it makes curdlan on culture midium.We'll explain curdlan on this page.
About
It is fermentative polysaccharide that microorganizm makes. It polymerizes two position;C₁and C₃on D-glucose. Its binding position is not straight. So it is cycloid structure.
File:Biwako-nagahama curdlan's-picture yoshiharu-otaki1.png
Property
It is insolubility in water. But it can become dispersing solution in cold water. And after high speed shuffle,the solution go to uniform dispersing solution. Moreover it become complete dispersing solution if it is mixxed by alkaline solution(avobe pH12). And last, dispersing solution become gel unnder the situation that it's heated by high temperature.
CrdS
セルC
Binary Vector
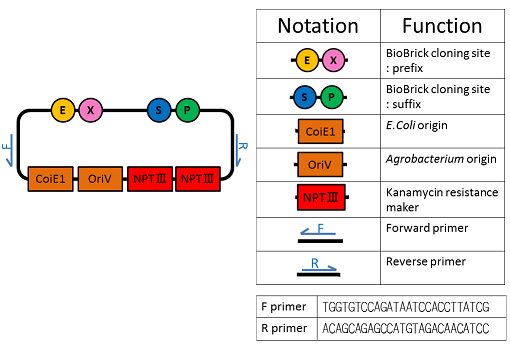
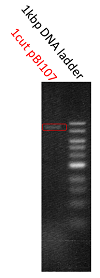
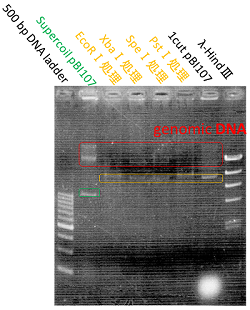
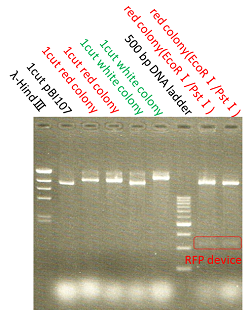
We named BBa_K1044006 "pBI107".This plasmid length is about 9,000 bp.
pBI107 is empty backbone vector which improved pBI333 which is an existing E.coli and a binary vector of Agrobacterium.It replicates E.coli and Agrobacterium, and can selection by kanamycin.
pBI107 was put in prefix and suffix of BioBrick's cloningsite.So, we can put a cassette in freely and pBI107 made possible expression of various BioBrick.
Moreover, OriV may be expression not only Agrobacterium but also various living things.
E.coli-ink
Another one of our project is E.coli ink.
We are planning to use Escherichia coli expressing the sperm whale myoglobin as ink. We are also trying to change the color of transgenic Escherichia coli by changing the growth and medium conditions.
Why do we use myoglobin as ink ?
Recently, fluorescent pigment is very famous and popular. It is nice to look at.
But, Many fluorescent pigment is only colouring when it’s exposed to ultraviolet rays.
We want to make pigment colouring under natural light. Thus,We decided to use myoglobin as ink.
Finally, We will draw my advisor’ face on Agrepaper with this E.coli ink !
The nucleotide sequence of myoglobin of sperm whale was already published, and We have synthesis company to synthesis the DNA fragment encoding the myoglobin.
Fig1 shows this DNA fragment sequence. Fig1 is published on paper.
Fig1.DNA fragment encoding myoglobin.
DNA synthesis company send us the DNA fragment with pTAKN-2. pTAKN-2 is plasmid that .Thus,We get DNA fragment with pTAKN-2.
Next,We insert to pUC18 to confirm the function of this DNA fragment.
We did double digestion pUC18 and pTAKN-2 with BamHⅠand SmaⅠ.This experiment protocol shows below.
This sample Incubated at 37℃ for 2h.add phenol chloroform mixture 50μℓ and Vortex. centrifugal at 4℃ with 13000rpm for 5min. pour supernatant into another tube. add 3M CH3COONa 5μℓ and 99.5% EtOH 125μℓ to supernatant and Vortex. centrifugal at 4℃ with 13000rpm for 10min. add mixture solution to pellet . mixture solution composition below.
This sample again incubated at 37℃ for 1h.add phenol chloroform mixture 50μℓ and Vortex. centrifugal at 4℃ with 13000rpm for 5min. pour supernatant into another tube. add 3M CH3COONa 5μℓ and 99.5% EtOH 125μℓ to supernatant and Vortex. centrifugal at 4℃ with 13000rpm for 10min. add 70% EtOH 500μℓ to pellet and up side down. Remove supernatant . dyr up at 65℃.add TEbuffer 10μℓ to pellet. Mix two samples and Ligation kit Ver2 . We use Ligation kit Ver2 on Ligation reaction.
Transformation (protocol shows なんちゃこんちゃら) Plating to LB-Amp plate.
Next day.
The Colony is on plate. We inoculate this colony into 2μℓ Amp in 2ml 2YT medium in culture tube
Next day
We did Miniprep of this sample.
Theoretical value is but this plasmid is supercoil structure. Therefore, Theoretical value is .
We confirm bund size is theoretical value with agarose electrophoresis.
We again inoculate this colony into 3μℓ 50ng/μℓ Amp in 30μℓ 100mM IPTG in 3ml 2YT medium in culture tube .
Next day, centrifugal this bacterial cell medium at 4℃ with 13000rpm for 1min.
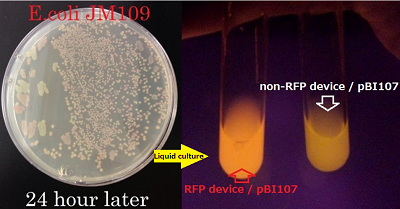
Fig2 shows part’s function!
Fig2.myoglobin is changed it’s color in Ecoli
left:pellet of JM109 with pUC18. right:pellet of JM109 with pUC18/Mb
Next,We use SDS/PAGE to confirm E.coli cloned pUC18/Mb produce myoglobin.
SDS/PAGE sample preparation protocol below.
Centrifugal Bacterial cells 200μℓ at 4℃ with 13000rpm for 1min. remove supernatant. Add 1×sample buffer to pellet and vortex. dip this sample into water bath at 98℃(not heat block). Let it sit on ice 30sec. Centrifugal the sample at 4℃ with 13000rpm for 10min.We use this sample 10μℓ for SDS/PAGE.
Fig3 shows Contative date of part’s function !
Fig3. SDS/PAGE analysis(CBB stain one super)with 15%gel
Lane1:JM109 with pUC18 Lane2:JM109 with pUC18/Mb
Next, We have to mutate this DNA fragment encoding myoglonbin because that includes EcoRⅠand XbaⅠsite. GAATTC that is EcoRⅠsite is changed to GAATTT. TTC is encoding amino acid and TTT too. As same CATT that is XbaⅠsite is changed to .
EcoRⅠsite pointmutation protocol below
First,We design the primer occurring pointmutation.Primer sequence below.
F1
R1
F2
R2
F3
R3
We use primer F1 and R2 to make the left end fragment on PCR. PCR product
We use primer F2 and R3 to make middle fragment on PCR.
We use thermal cycler on PCR. but,middle fragment theoretical value is about 60 base pair, it is very short. So this PCR don’t amplify DNA fragment we targeting but plasmid itself. PCR enzyme using by us is Prime star Max. Prime star Max is highly efficient enzyme. We replace Prime star Max with prime star hot start , but result is same.
Hereupon we hit upon an idea. We worked PCR by hand with water bath at 55℃, 70℃ and 98℃ because We think thermal cycler appear gradient temperature and this is a cause of failure.. We dip sample in water bath one by one. 98℃,55℃,70℃,98℃,55℃・・・・.
 "
"