Team:UChicago/Plan
From 2013.igem.org
(→Constructing the kerA BioBricks) |
|||
| Line 34: | Line 34: | ||
Once we carried out Gibson assembly, we amplified the kerA sequence with PCR. We ran our PCR product in a gel and assumed that the right sequence was obtained based on the size of the DNA band. Once we had a lot of kerA DNA to work with, we decided it would be best to insert the BioBrick into an Amp resistant backbone provided in the iGEM kit. To do so, we digested the PCR product and the backbone with EcoRI and PstI. We set up a ligation and transformed the plasmid into competent E. coli DH5-alpha. Nonetheless, we were unable to get any transformants. Prior to the transformation with the plasmid containing kerA, we tested the transformation efficiency of our competent cells so we discarded the possibility that were doing the transformations incorrectly. Therefore, we thought that maybe the problem was with the assembly, PCR, digestion, or ligation of kerA. We tried using different conditions (temperatures, incubation time, and different enzymes) to eliminate our sources of error, but we were unable to get any transformants. | Once we carried out Gibson assembly, we amplified the kerA sequence with PCR. We ran our PCR product in a gel and assumed that the right sequence was obtained based on the size of the DNA band. Once we had a lot of kerA DNA to work with, we decided it would be best to insert the BioBrick into an Amp resistant backbone provided in the iGEM kit. To do so, we digested the PCR product and the backbone with EcoRI and PstI. We set up a ligation and transformed the plasmid into competent E. coli DH5-alpha. Nonetheless, we were unable to get any transformants. Prior to the transformation with the plasmid containing kerA, we tested the transformation efficiency of our competent cells so we discarded the possibility that were doing the transformations incorrectly. Therefore, we thought that maybe the problem was with the assembly, PCR, digestion, or ligation of kerA. We tried using different conditions (temperatures, incubation time, and different enzymes) to eliminate our sources of error, but we were unable to get any transformants. | ||
If everything had worked out, we would have carried on to alter the kerA BioBrick we first assembled so that it contained a his tag. | If everything had worked out, we would have carried on to alter the kerA BioBrick we first assembled so that it contained a his tag. | ||
| + | |||
Orange: prefix | Orange: prefix | ||
| + | |||
Green: suffix | Green: suffix | ||
| + | |||
Purple: kerA signal peptide | Purple: kerA signal peptide | ||
| Line 41: | Line 44: | ||
[[File:kerA variant B.jpg]] | [[File:kerA variant B.jpg]] | ||
[[File:kerA variant C.jpg]] | [[File:kerA variant C.jpg]] | ||
| - | |||
== Construct pUB110 backbone == | == Construct pUB110 backbone == | ||
Revision as of 22:11, 27 September 2013
Contents |
Summer Plan and Challenges
Constructing the kerA BioBricks
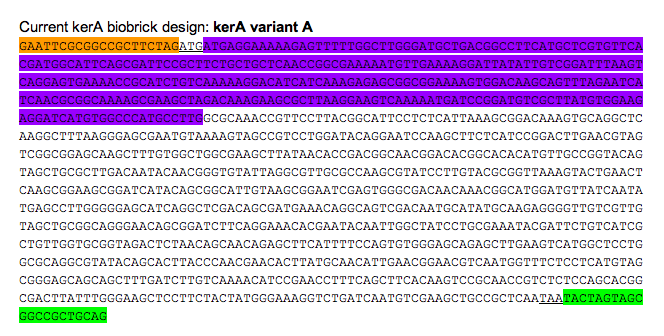
It was less costly to order gBlocks than purchase the Bacillus licheniformis PWD-1 strain that has the kerA gene, thus, we opted to assemble to kerA gene with gBlocks from IDT. Once we carried out Gibson assembly, we amplified the kerA sequence with PCR. We ran our PCR product in a gel and assumed that the right sequence was obtained based on the size of the DNA band. Once we had a lot of kerA DNA to work with, we decided it would be best to insert the BioBrick into an Amp resistant backbone provided in the iGEM kit. To do so, we digested the PCR product and the backbone with EcoRI and PstI. We set up a ligation and transformed the plasmid into competent E. coli DH5-alpha. Nonetheless, we were unable to get any transformants. Prior to the transformation with the plasmid containing kerA, we tested the transformation efficiency of our competent cells so we discarded the possibility that were doing the transformations incorrectly. Therefore, we thought that maybe the problem was with the assembly, PCR, digestion, or ligation of kerA. We tried using different conditions (temperatures, incubation time, and different enzymes) to eliminate our sources of error, but we were unable to get any transformants. If everything had worked out, we would have carried on to alter the kerA BioBrick we first assembled so that it contained a his tag.
Orange: prefix
Green: suffix
Purple: kerA signal peptide
Construct pUB110 backbone
In order to express kerA in B. subtilis in high amounts, we need to use a high copy number plasmid. Since high copy number plasmids for B. subtilis are not available form the iGEM HQ, we decided to design our own. We derived our design from pUB110 because it has been widely used in B. subtilis.
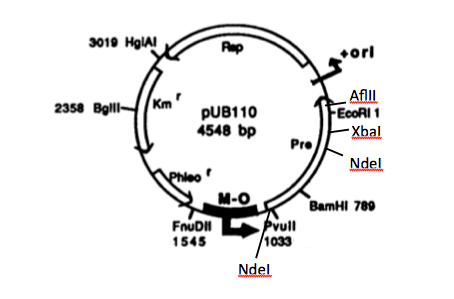
Source: Boe, L. A. R. S., Gros, M. F., te Riele, H. E. I. N., Ehrlich, S. D., & Gruss, A. (1989). Replication origins of single-stranded-DNA plasmid pUB110. Journal of bacteriology, 171(6), 3366-3372.
In the image, M-O represents the origin replication that is recognized by B. subtilis, while ori+ is the origin of replication used by S. aureus. Because this vector includes EcoRI and XbaI sites, we had to find a way to remove them before we turned the vector into a BioBrick backbone. We chose to digest pUB110 with NdeI and AflII, which would remove the Pre gene along with the EcoRI and XbaI sites from the vector. However, this gene is not essential for replication in B. subtilis. We then ran the digest in a gel and did a gel extraction to isolate the part of the vector that we needed.
In order to add in the BioBrick prefix and suffix to the vector, we designed a linker that was synthesized as two separate oligos. The DNA sequence of the two oligos are as follows:
5' attcgtcttaaggaattcgcggccgcttctagagtactagtagcggccgctgcagcatatgtcatac 3'
5' gtatgacatatgctgcagcggccgctactagtactctagaagcggccgcgaattccttaagacgaat 3'
We annealed the two oligos in duplex buffer (recipe from IDT) to use the duplex as a linker. Once the oligos were annealed, the linker was digested with NdeI and AflII. We then set up an overnight ligation of the digested, gel purified pUB110 and digested pUB110 linker, which would ideally lead to the formation of a backbone compatible with BioBricks. Nonetheless, when we were unable to get any transformants with our modified vector. To make sure that we weren't doing anything wrong with the transformation procedure of B. subtilis, we tried to transform the original pUB110. The transformation was successful, which led us to think that the problem may be with our modified vector. The failure of transformations could have been attributed to incomplete digestions, too little DNA retrieved from the gel extraction, or incomplete ligation. We attempted to address these potential problems by using new AflII and NdeI restriction enzymes with varying incubation times, different kits for gel extractions, and overnight ligations. Despite all our attempts, we did not successfully transformed B. subtilis with our modified backbone.
If our transformation of the modified pUB110 vector had worked, we would have picked transformants, grown them overnight to amplify the quantity of backbone DNA, and then done a miniprep to isolate the modified pUB110 backbone. We would have then sent the vector for sequencing. Once we verified that the backbone sequence were correct, we would have tested expression of the backbone. Using the NEB assembly kit, we would have digested an upstream part or promoter, a RFP BioBrick, and the modified pUB110 backbone. Once the construct were assembled, we would have transformed it into B. subtilis and checked for RFP expression.
Do the following 3 step assemblies:
For E. coli BL21-De3
- Constitutive expression cassette (BBa_K314100: plate 1, well 1G chloramphenicol res vector) + KerA variant C (in amp res vector) + kan res biobrick backbone
- Lac induced expression cassette (BBa_K314103: plate 1, well 4D chloramphenicol res vector) + KerA variant C (in amp res vector) + kan res biobrick backbone
For B. subtilis
- Constitutive promoter Pveg +RBS spoVG + SacB (BBa_K541501:
plate 1, well 11E chloramphenicol res vector) + KerA variant C (in amp res vector) + kan res biobrick backbone
- Constitutive promoter Pveg + RBS spoVG (BBa_K143053: 2012 plate 3, well 20A kan and amp res vector) + KerA variant B (in chloramphenicol res vector) + kan res biobrick backbone
Pick transformants of each, do miniprep and send them for sequencing
Test keratinase expression and activity in E. coli BL21-De3 kerA transformants
Purchase commercially available keratinase A to compare activity to our keratinase Potential sources from which to purchase (since Sigma is backordered): http://www.biocat.com/products/PBL01.1.02-PR http://www.interchim.eu/reponse_cch.php?ref=520&manufacturer=0&search=keratinase&search2= http://www.interchim.fr/ft/J/JV4050.pdf http://www.proteosbiotech.com/pdfs/pure_keratinase_100_en.pdf http://www.proteosbiotech.com/index.php/en/reactivos-de-laboratorio-en/keratinasa/pure-100-keratinase-detail
Or purchase B. licheniformis PWD-1 strain.
Detect keratinase expression in cell lysate of E. coli BL21-De3 kerA transformants (constitutive and inducible) by western blot with antibody against his tag.
If expression is successful, do a cell lysis of E. coli BL21-De3 kerA transformants and test keratin azure assay.
Also, KerA could be isolated with nickel column (because kerA is his tagged).
Test keratinase expression and activity in B. subtilis kerA transformants
Grow B. subtilis kerA transformants on keratin plates (made with LB agar [nutrient rich] + keratin from DMSO dissolved feathers). Grow WT/non-transformed B. subtilis on keratin plate. Determine if there is any non-specific protease activity. If there is a clearing around the non-transformed B. subtilis controls, this is not a good way to determine keratinase expression. Compare the clearing of keratin plate accomplished by kerA transformants and WT/non-transformants. If possible, compare activity to B. licheniformis PWD-1 strain.
Grow B. subtilis kerA transformants and from a cell lysis, do western blot to detect kerA his tag expression.
After growing B. subtilis kerA transformants O. N. (unsure for how long) in liquid medium (LB broth plus antibiotic), spin down cells and store supernatant/media. Do western blot on media to detect kerA-his secretion.
Also, test keratin azure assay on supernatant from O. N. culture to determine keratinase activity.
Improve KerA activity by mutagenesis
Use Taq polymerase to introduce mutations in kerA. Transform mutated kerA into B. subtilis and plate on keratin plates. Detect clearing area around transformants. Do keratin azure assay.
 "
"