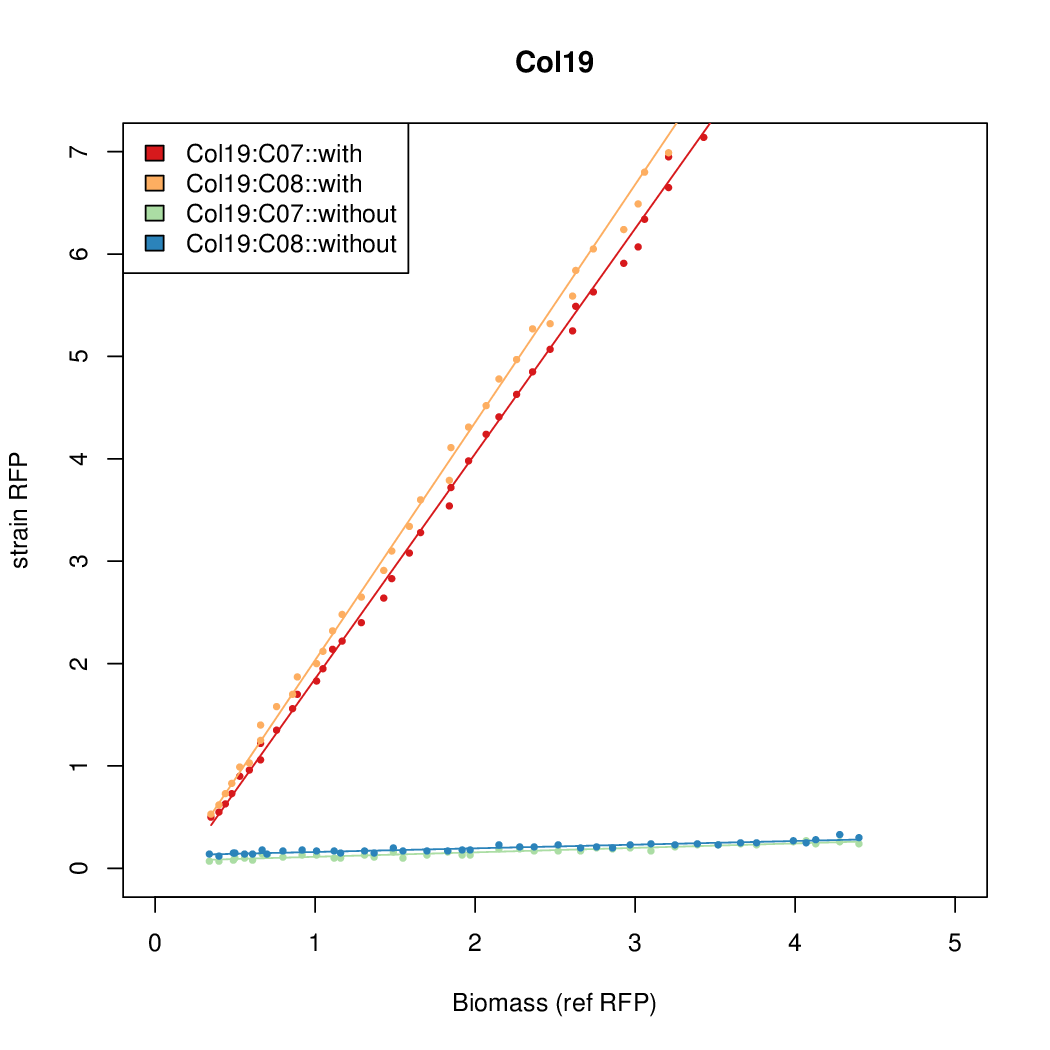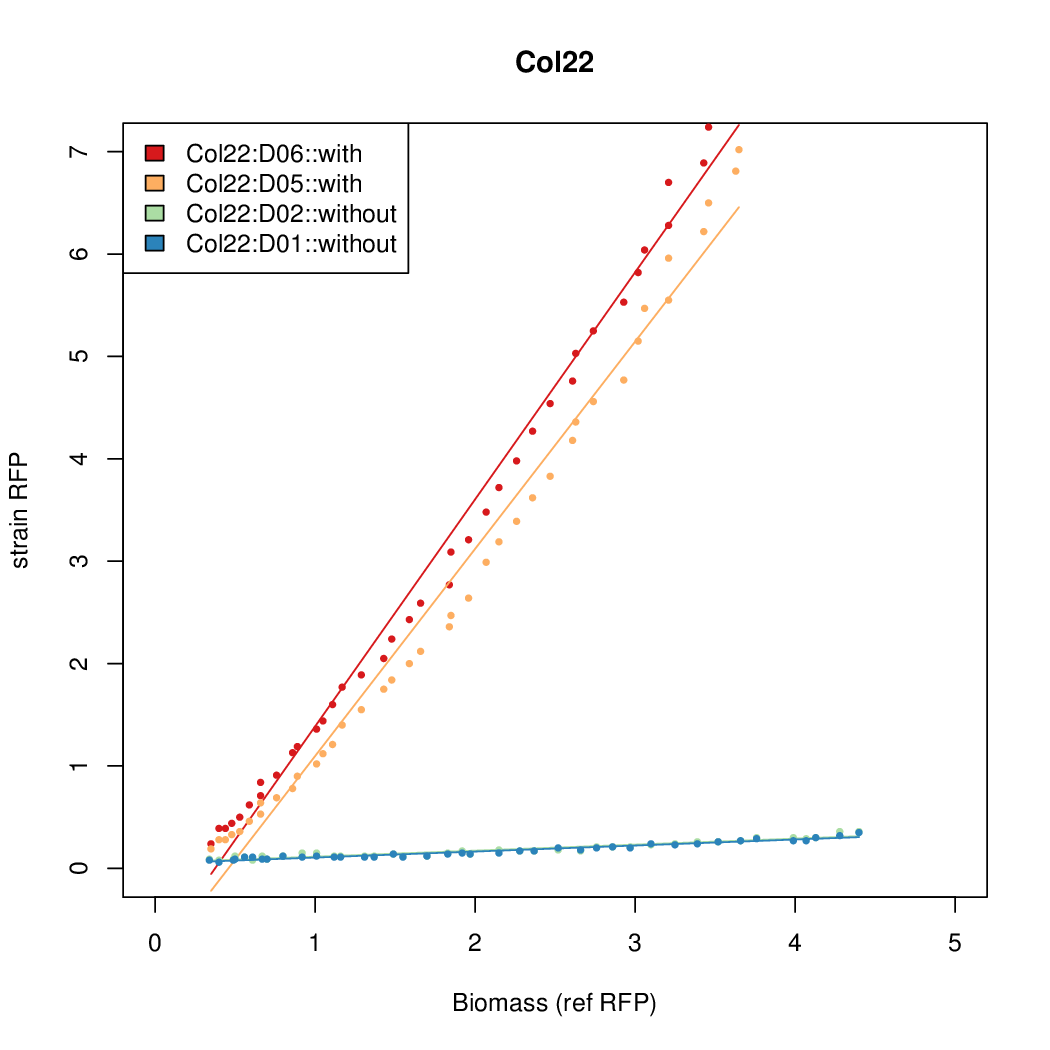Team:DTU-Denmark/pBAD SPL
From 2013.igem.org
(Difference between revisions)
(→Data analysis) |
(→Details) |
||
| Line 32: | Line 32: | ||
=== Details === | === Details === | ||
| + | |||
| + | {| | ||
| + | !with.1 | ||
| + | !with.2 | ||
| + | !without.1 | ||
| + | !without.2 | ||
| + | |- | ||
| + | |Col2||8.9025||7.8699||0.727||0.9552 | ||
| + | |- | ||
| + | |Col3||9.2724||12.1142||0.5248||0.6982 | ||
| + | |- | ||
| + | |Col4||9.4571||11.4522||0.5231||0.2508 | ||
| + | |- | ||
| + | |Col12||6.3641||10.5389||0.5897||0.6869 | ||
| + | |- | ||
| + | |Col10||7.9697||9.7949||0.3392||0.733 | ||
| + | |- | ||
| + | |Col9||7.8563||20.1094||0.6995||0.7432 | ||
| + | |- | ||
| + | |Col8||12.2318||15.4548||0.4203||0.4538 | ||
| + | |- | ||
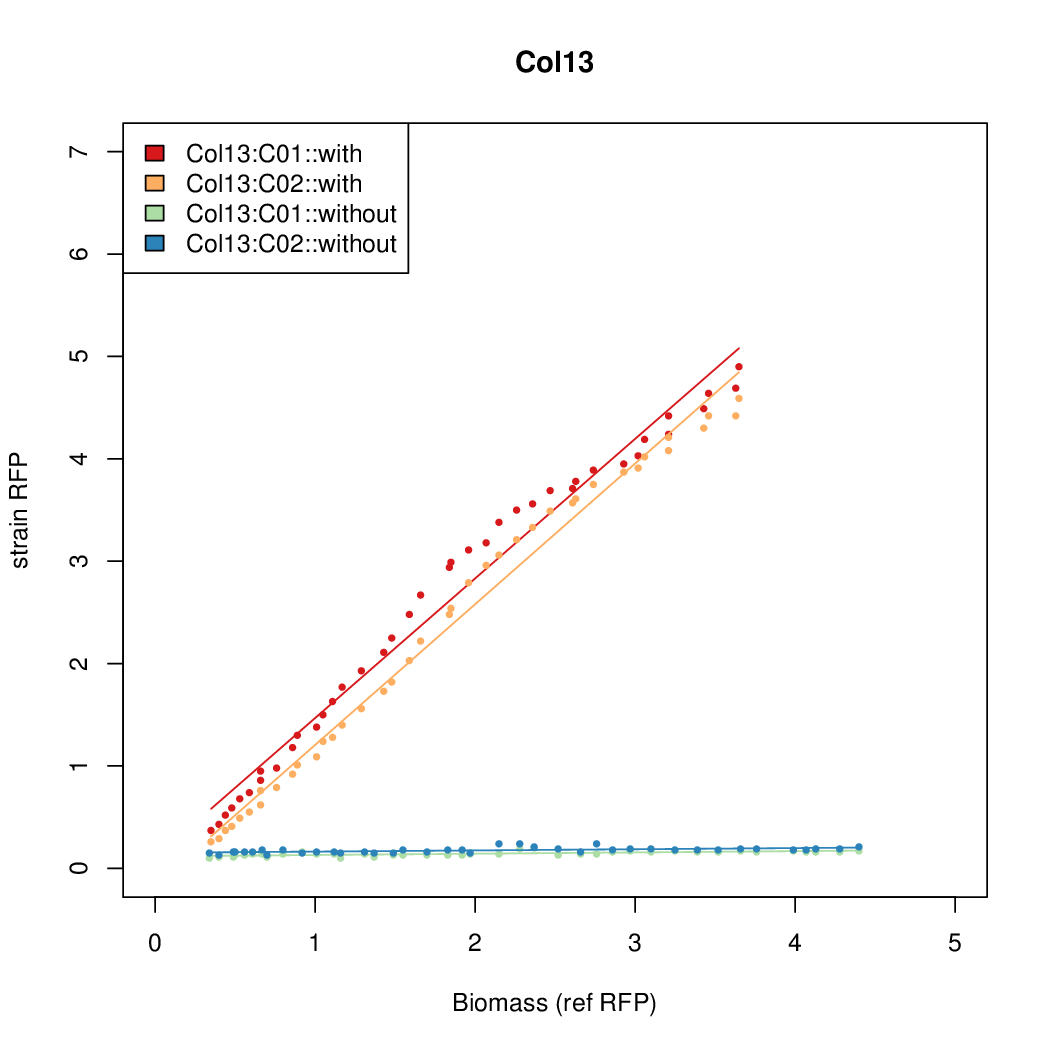
| + | |Col13||11.0377||7.3343||0.482||0.4641 | ||
| + | |- | ||
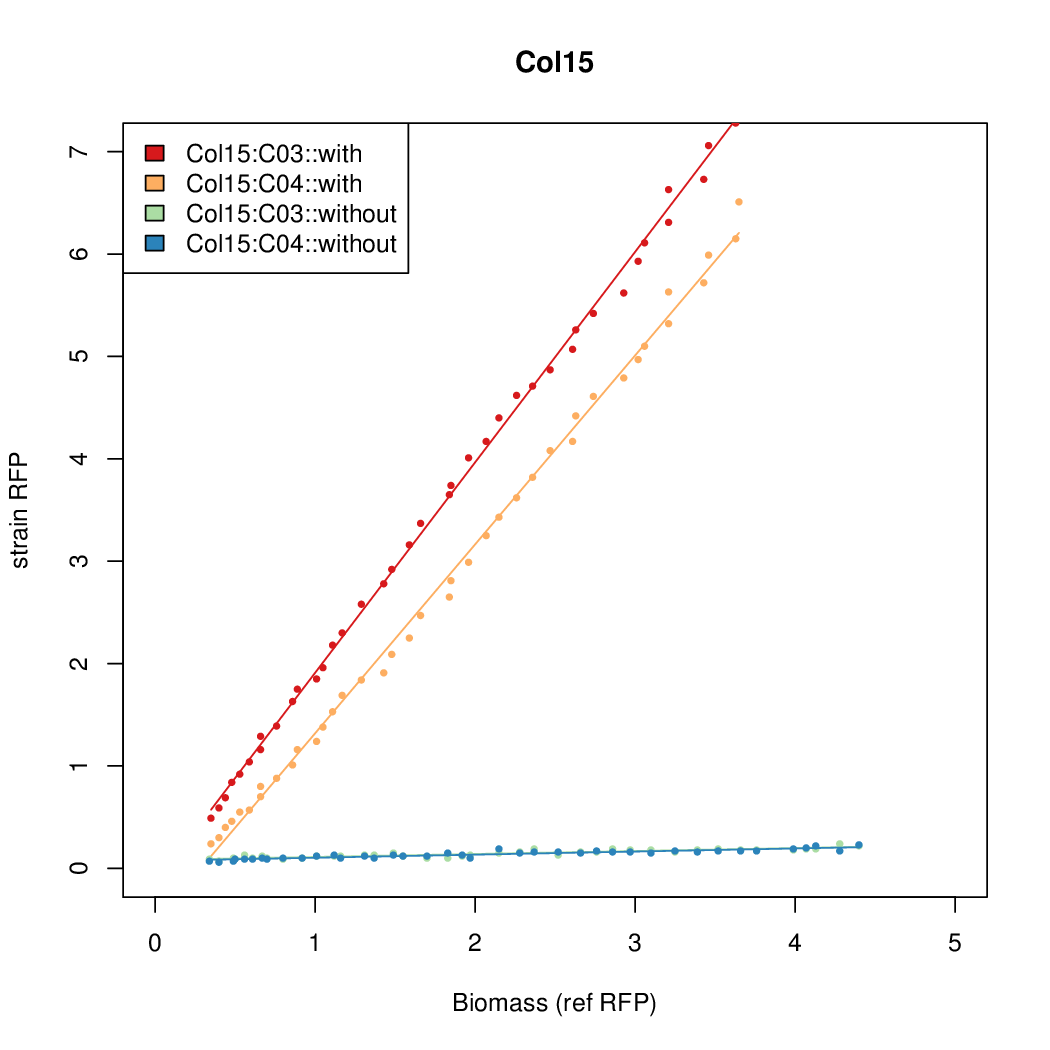
| + | |Col15||15.6817||8.2707||0.8169||0.1343 | ||
| + | |- | ||
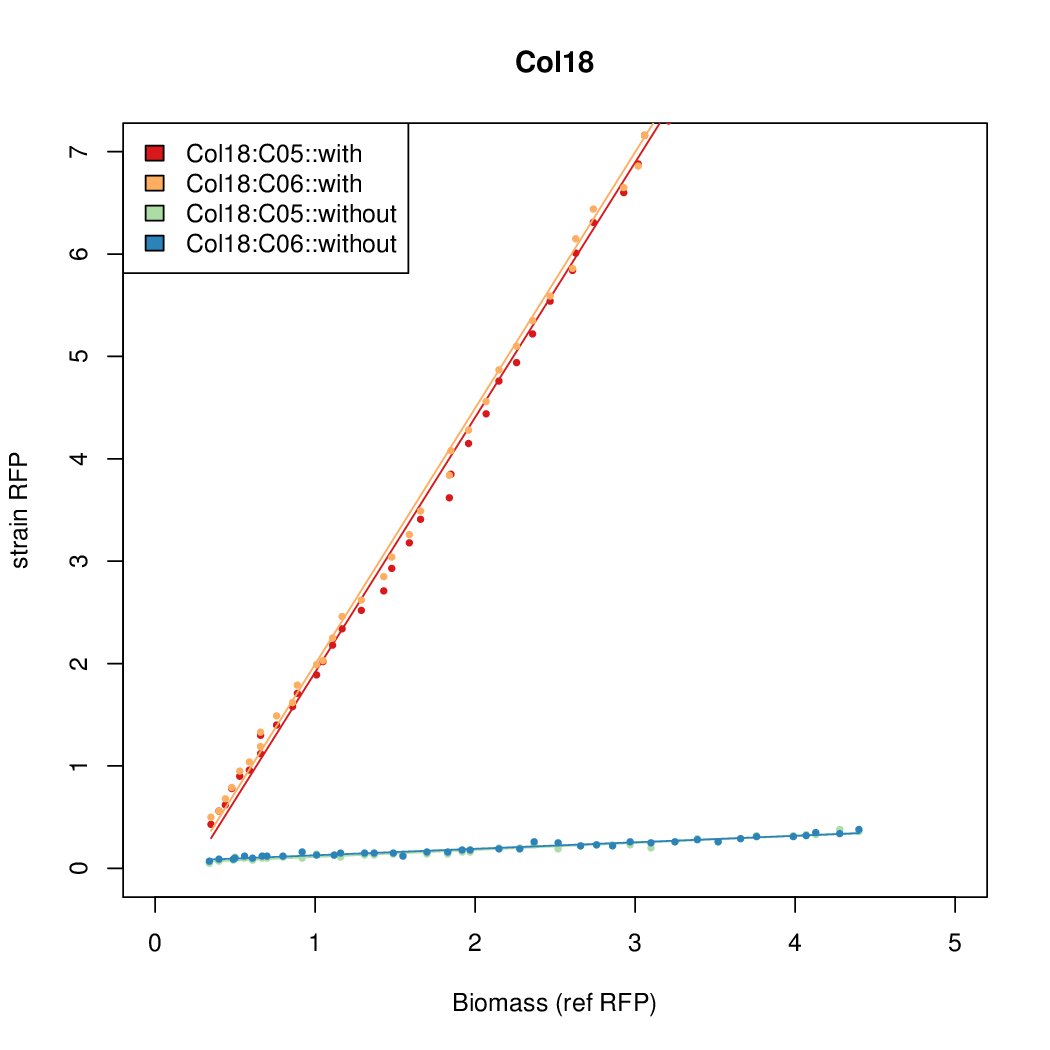
| + | |Col18||14.7916||15.5736||0.6674||0.6745 | ||
| + | |- | ||
| + | |Col19||14.2126||16.4898||0.4545||0.3566 | ||
| + | |- | ||
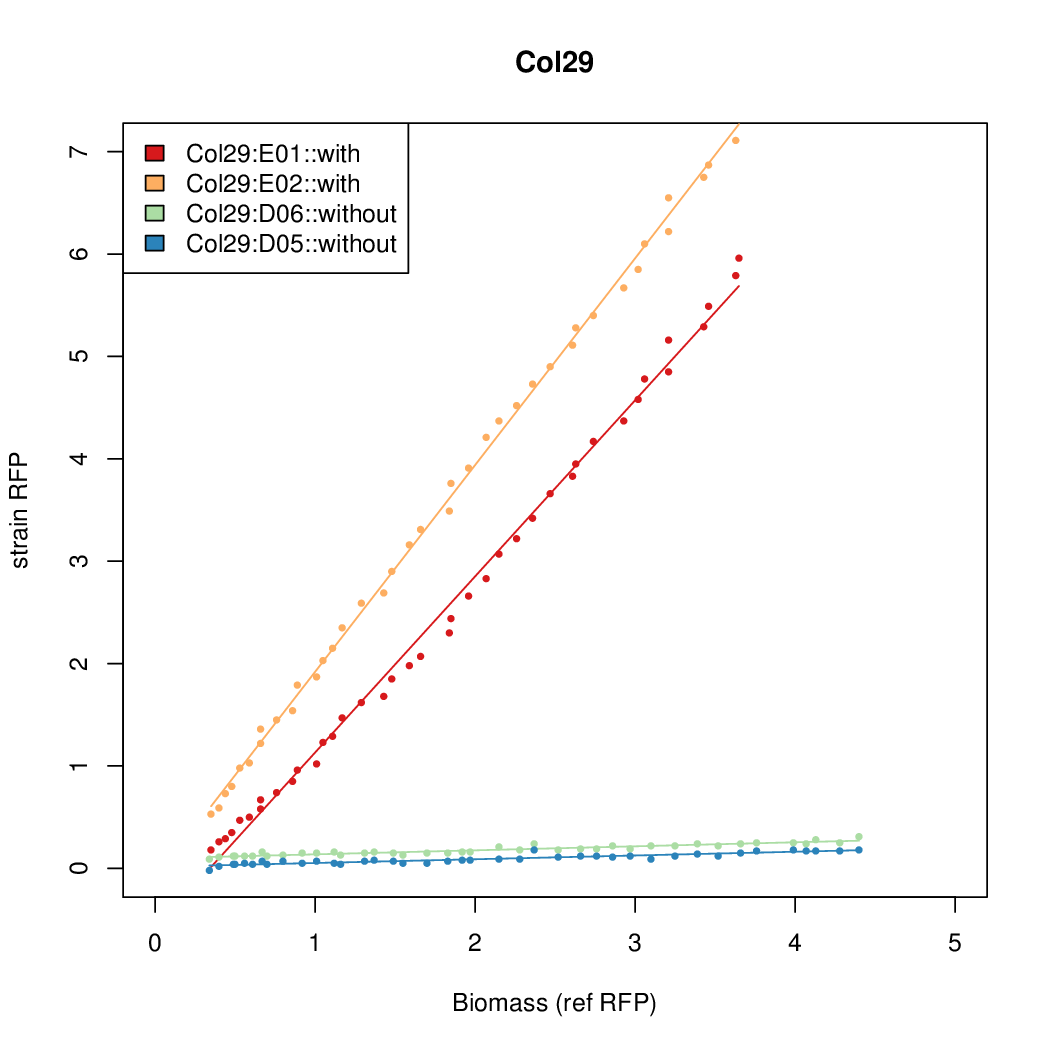
| + | |Col29||7.1853||16.3467||0.5445||0.5013 | ||
| + | |- | ||
| + | |Col26||9.7724||9.6269||0.7118||0.7865 | ||
| + | |- | ||
| + | |Col22||8.4168||5.5958||0.6049||0.5645 | ||
| + | |- | ||
| + | |Col33||9.1982||8.9987||0.6508||1.374 | ||
| + | |- | ||
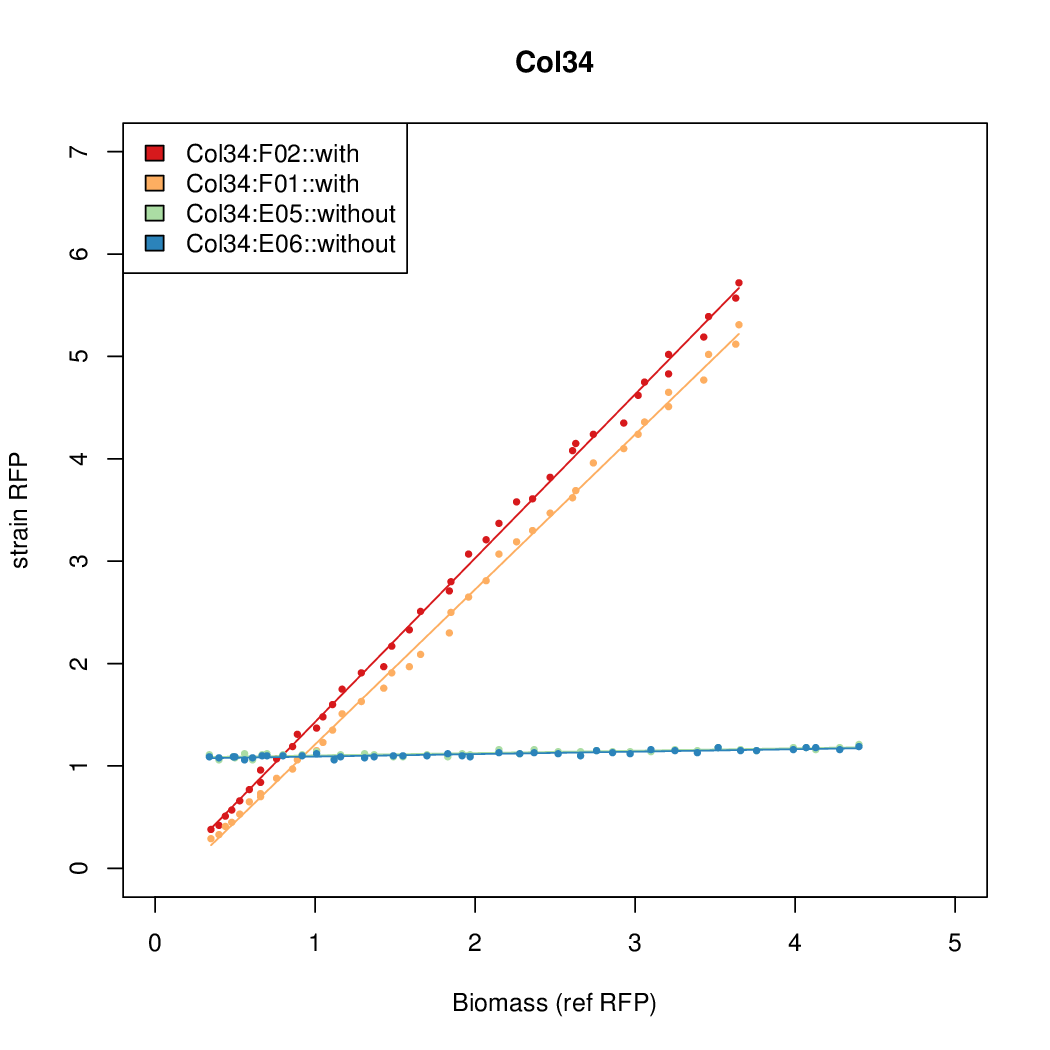
| + | |Col34||10.6987||7.883||0.5067||0.5031 | ||
| + | |- | ||
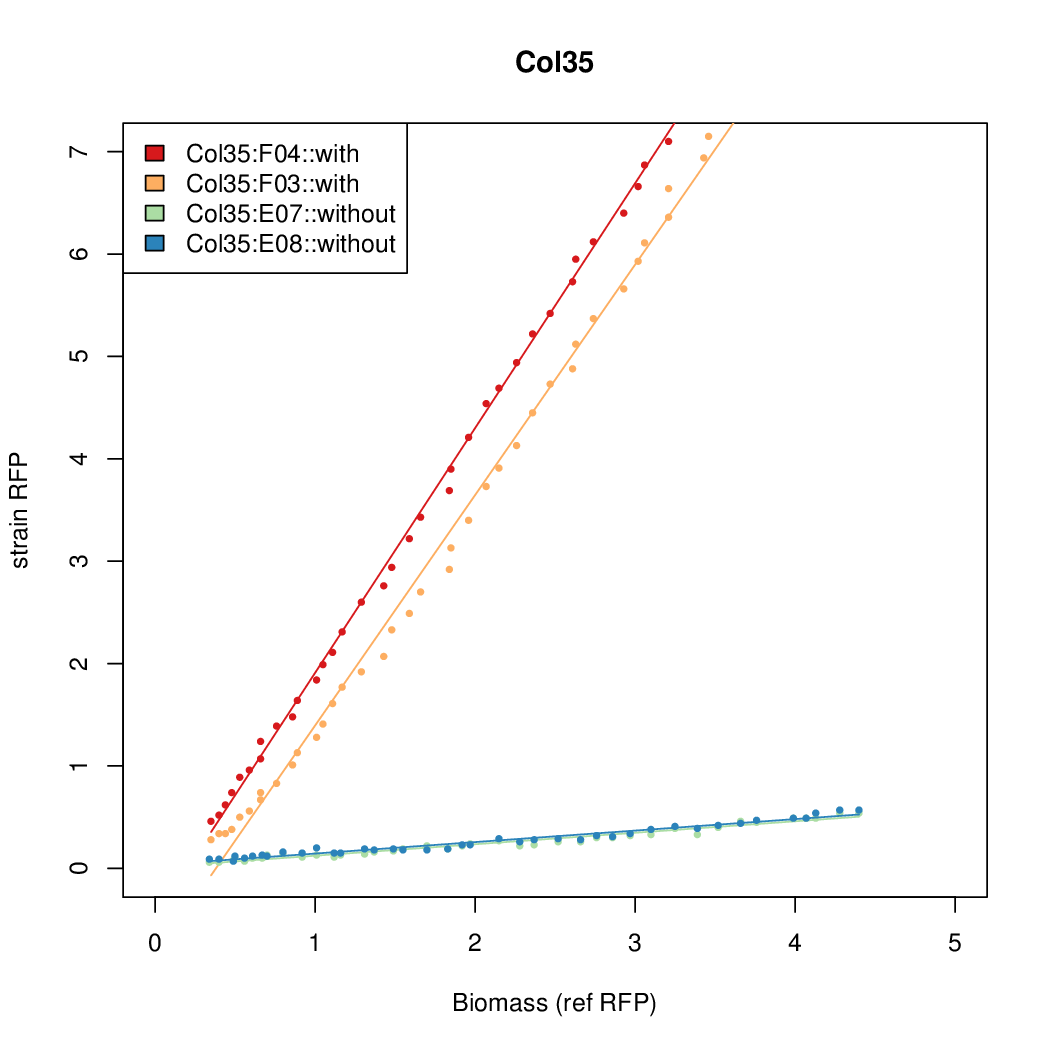
| + | |Col35||13.8427||7.5469||0.4363||0.6281 | ||
| + | |- | ||
| + | |ConRef||6.506||7.9323||8.7811||7.9323 | ||
| + | |} | ||
[[File:dtu-Fss-plot-col2.png|300px]] | [[File:dtu-Fss-plot-col2.png|300px]] | ||
Revision as of 08:21, 1 October 2013
pBAD SPL
Contents |
pBAD synthetic promoter library
As a tool for expressing lethal proteins in E. coli we made a synthetic promoter library (SPL, [http://dspace.mit.edu/handle/1721.1/60080 RFC 63]) with the pBAD arabinose inducible promoter. The concept was taken from the DTU iGEM team from 2010.
Methods
Experimental procedure
- Random promoter sequences were ordered matching the sequence CTGACGNNNNNNNNNNNNNNNNNNTAWWATNNNNA.
- USER cloning to add RFP downstream of promoter.
- Colonies were plated.
- Colonies that were not red prior to induction with arabinose but that did turn red after induction with arabinose were selected.
- Wells were inoculated from overnight cultures of each of the selected colonies
- One plate was run with arabinose, and another plate was run without arabinose.
Data analysis
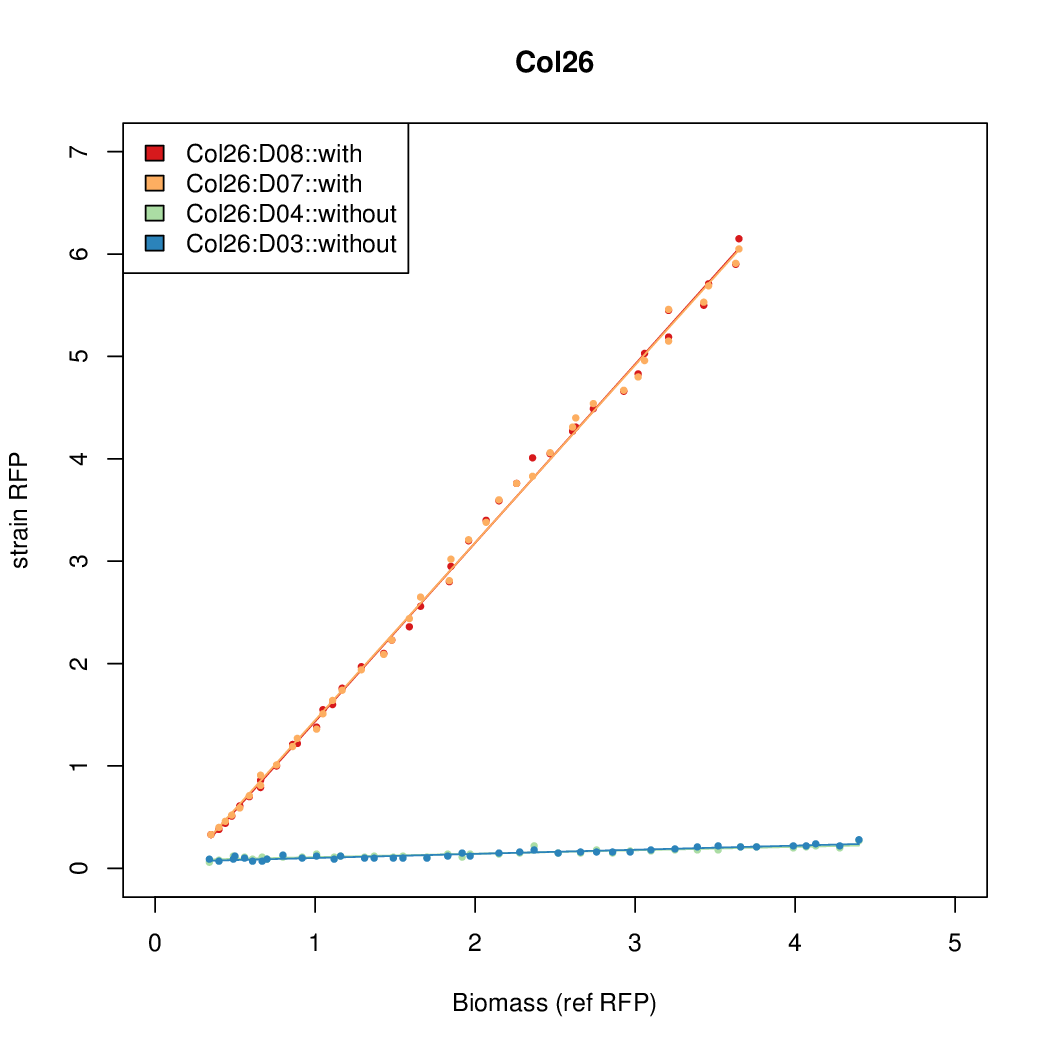
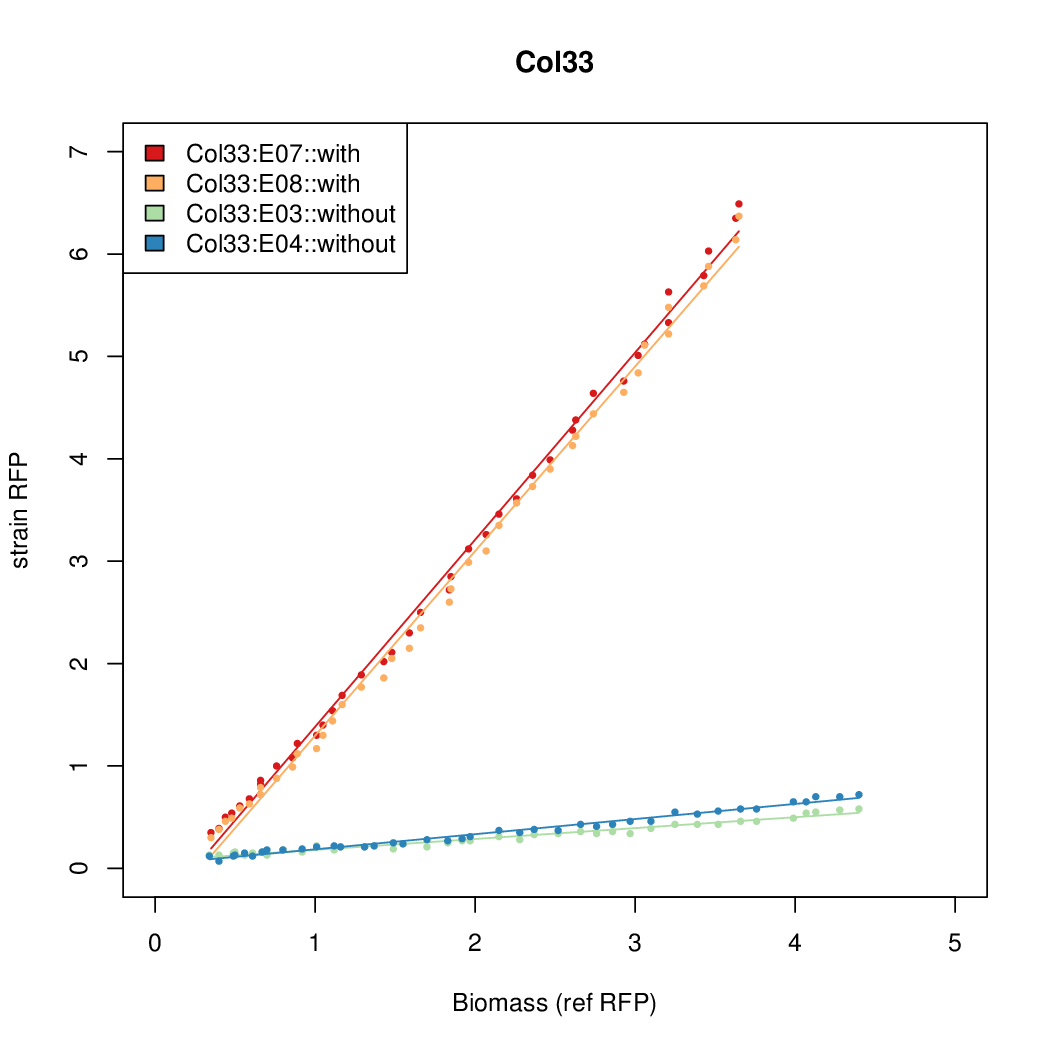
- Data was collected from the Biolector, and analyzed using a series of R scripts written by Chris Workman (unpublished).
- The maturation and degradation times for mCherry were both assumed to be 40 min. TODOref
- The growth rate, mu, was estimated to be 1.28 (from an average of all wells on all plates) since we expect each strain to grow at the same rate.
- A time window representing exponential growth was selected (between 1 and 4.5 hours).
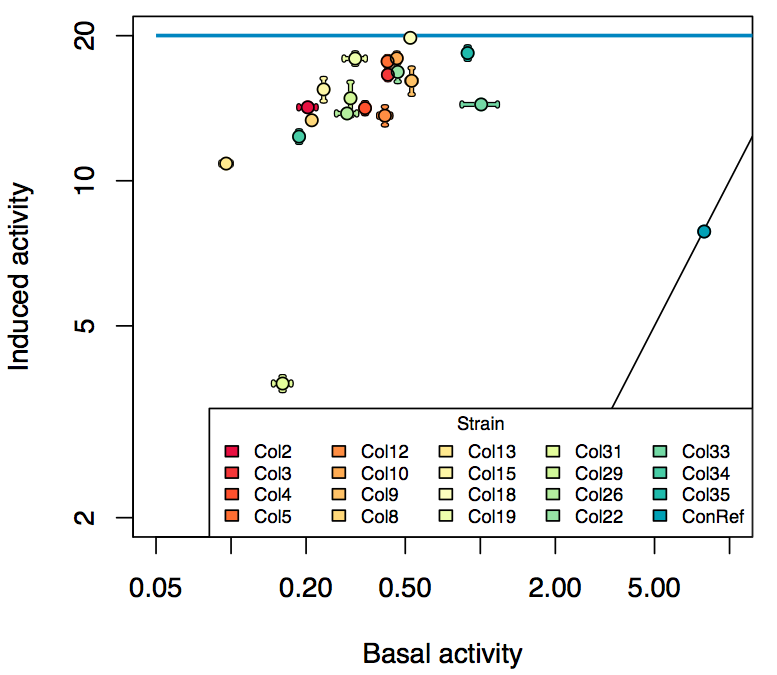
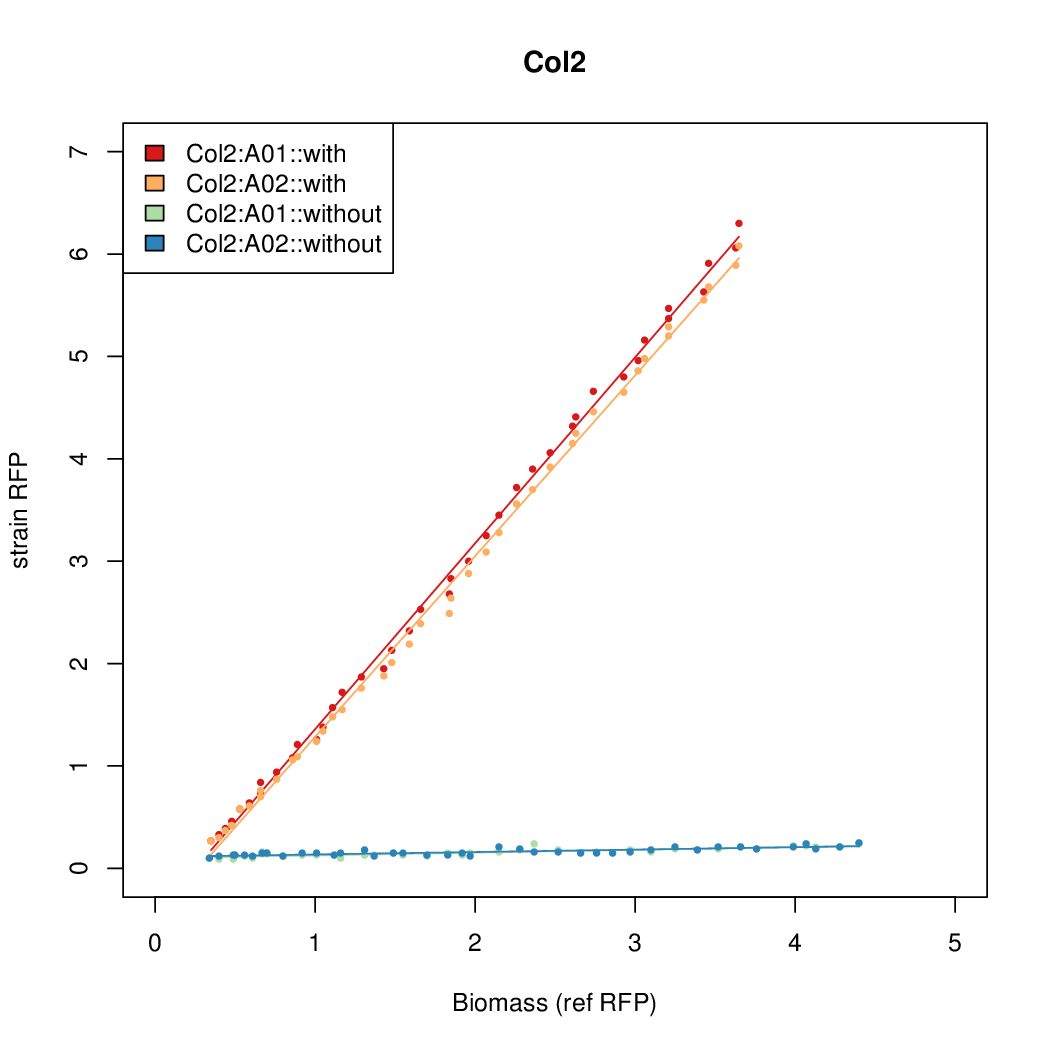
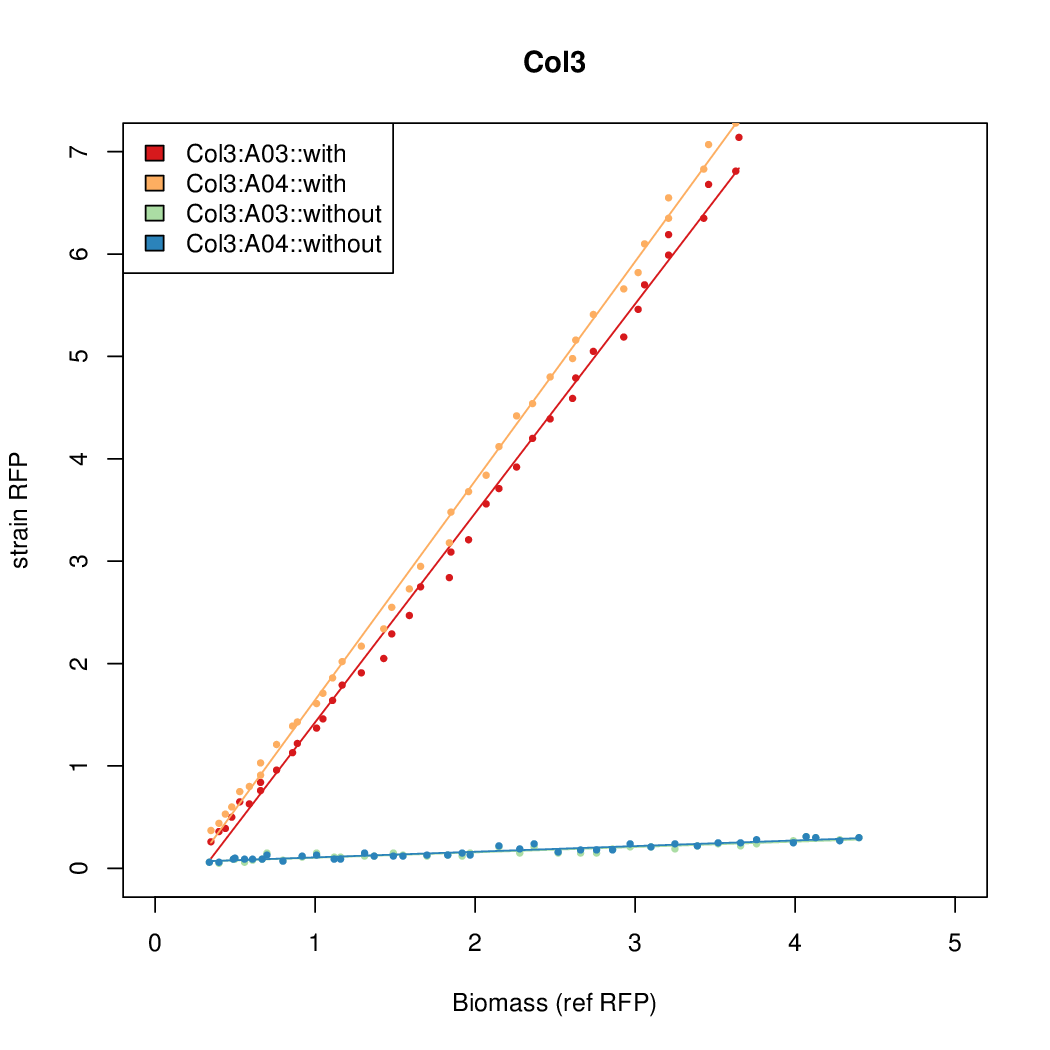
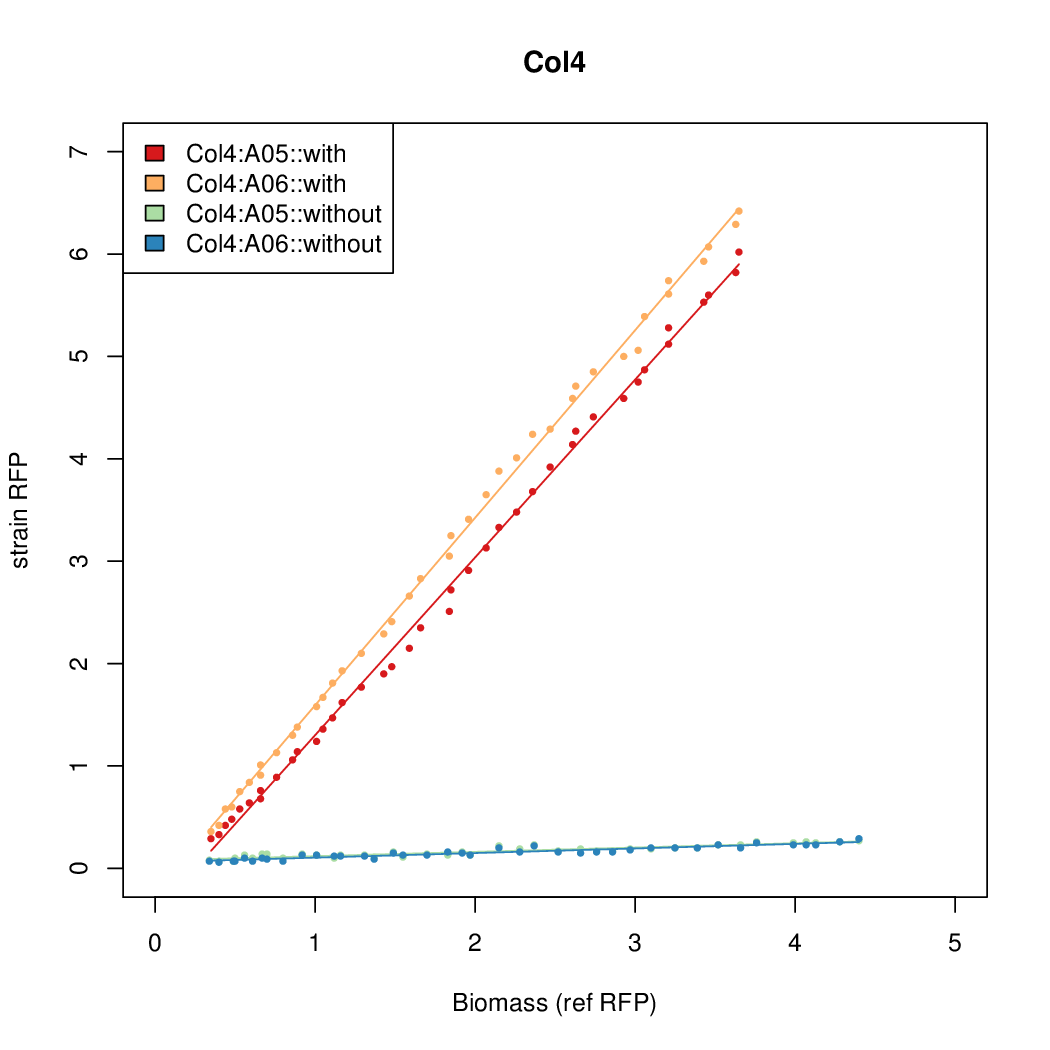
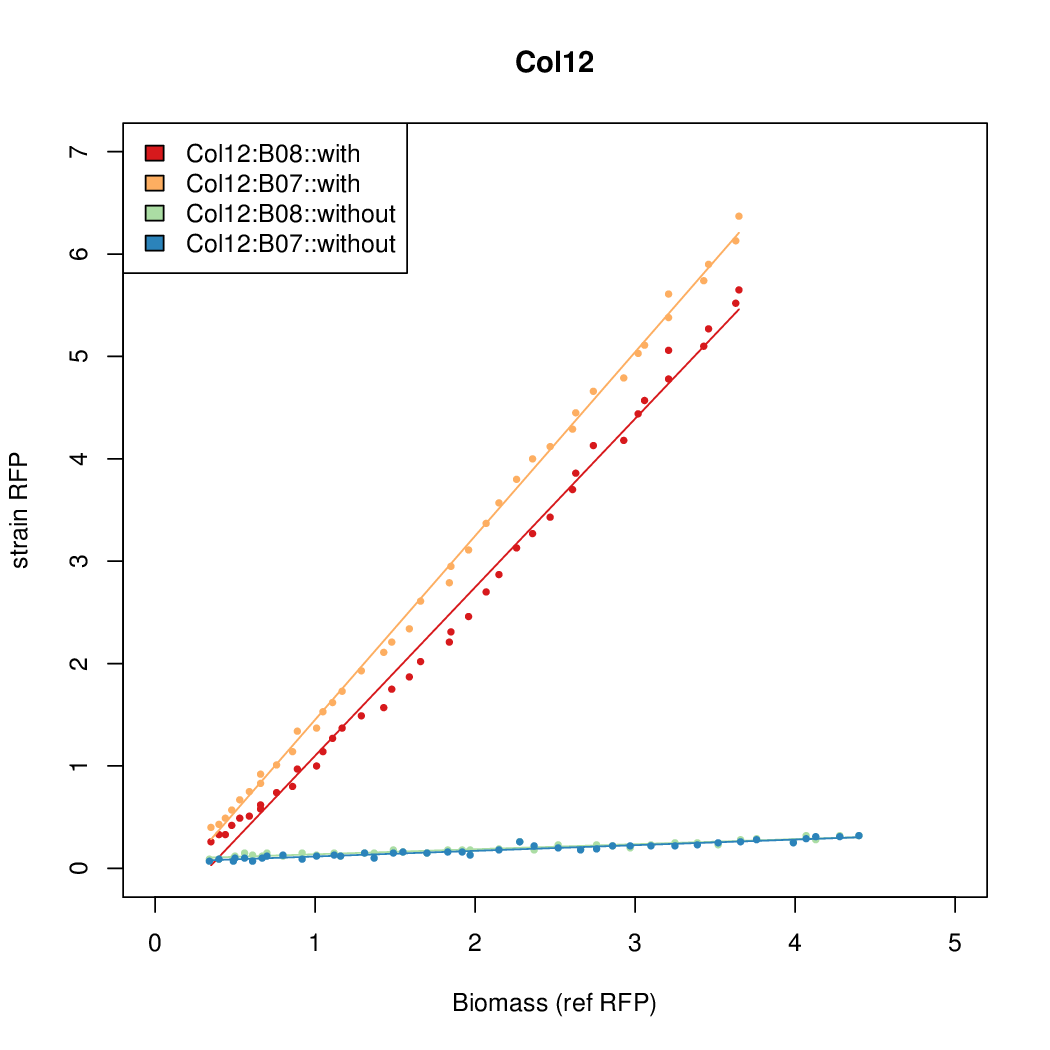
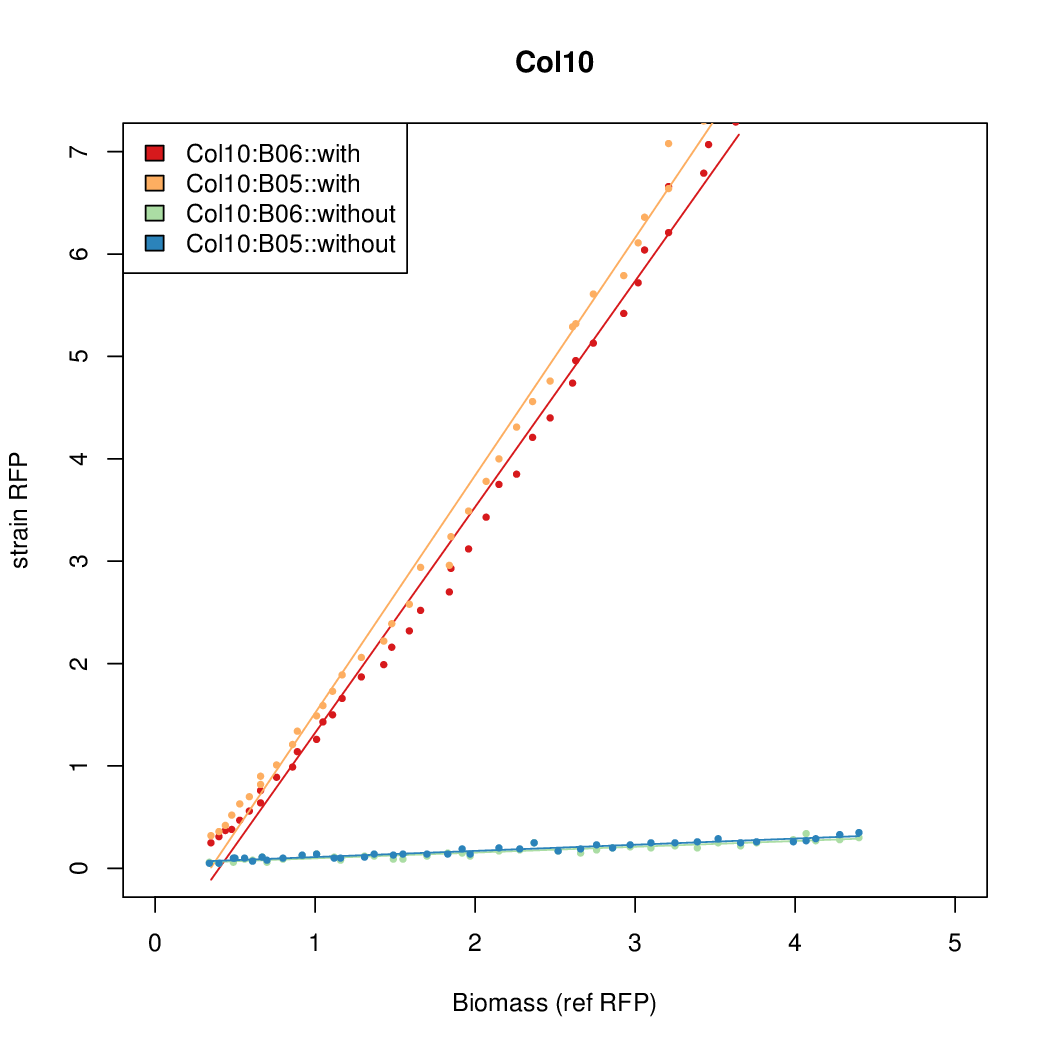
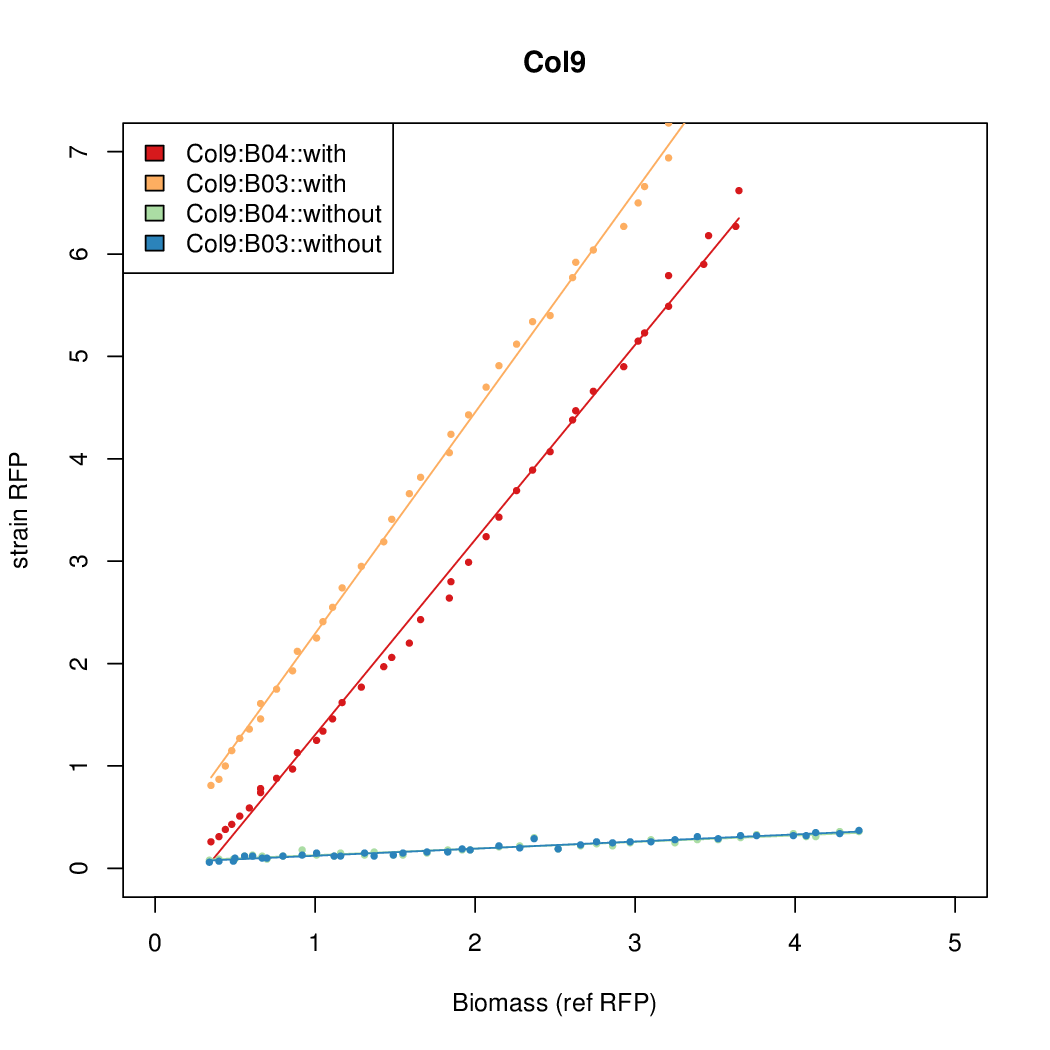
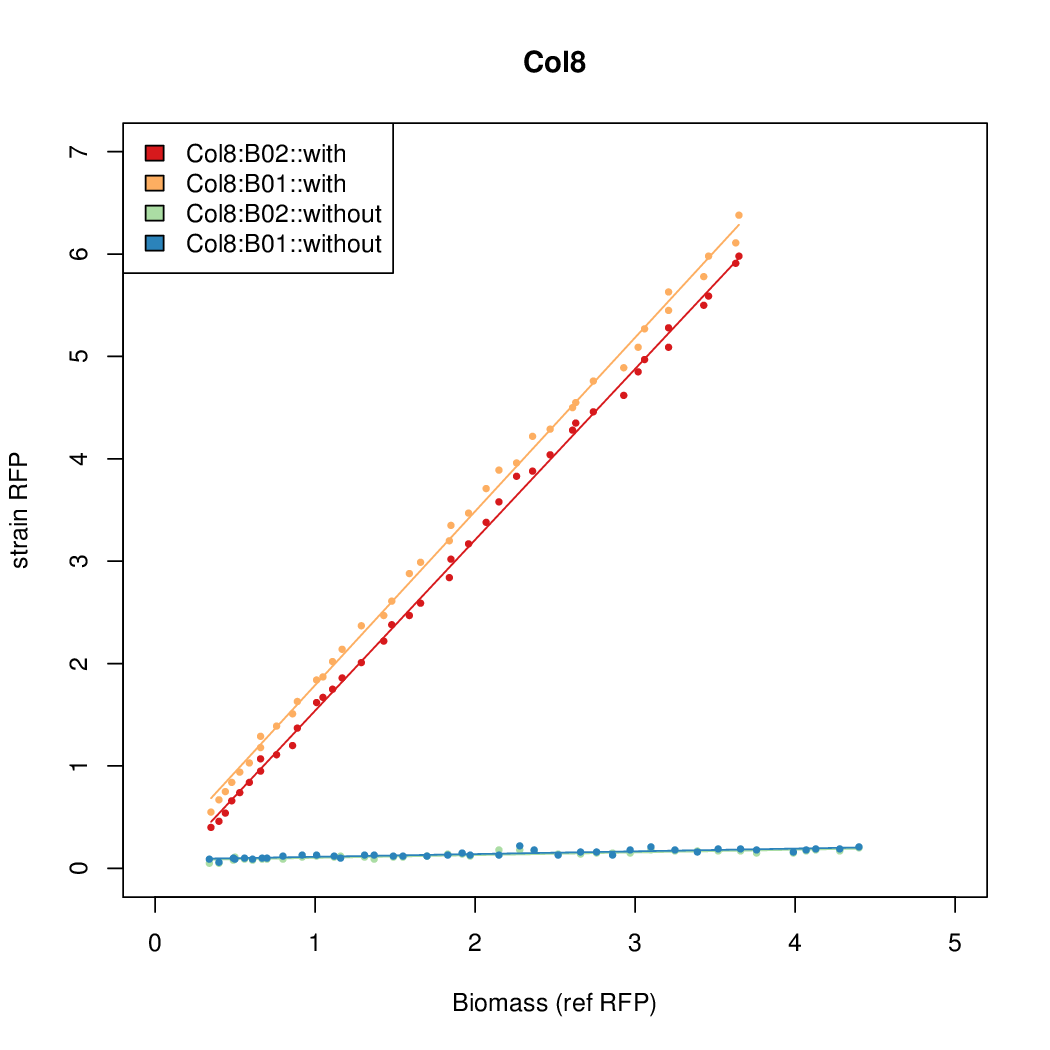
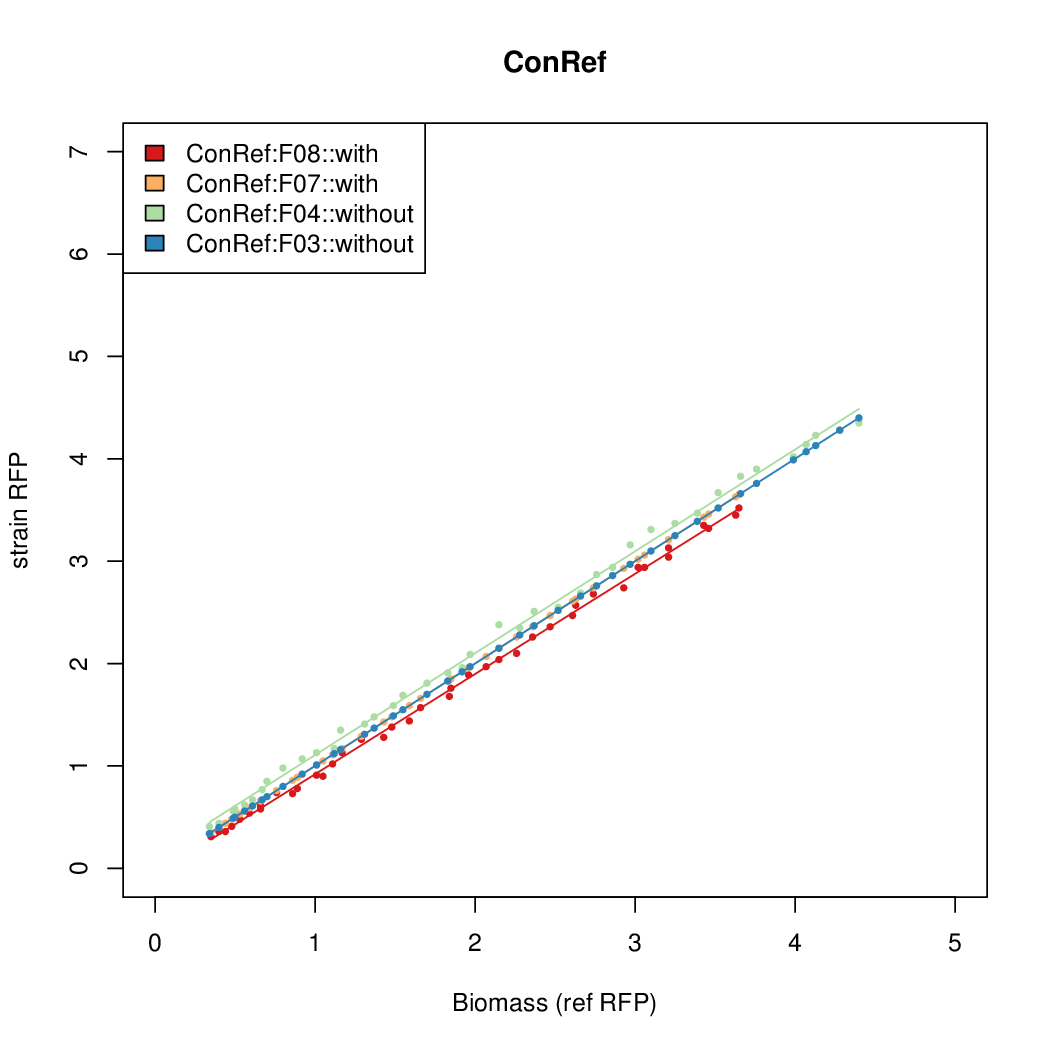
- The RFP measurement for a constitutively expressed strain was used as a standard measure of growth. This is plotted on the x-axis in the detailed plots per colony below.
- Figures were plotted using R.
Results
Summary
Details
| with.1 | with.2 | without.1 | without.2 | |
|---|---|---|---|---|
| Col2 | 8.9025 | 7.8699 | 0.727 | 0.9552 |
| Col3 | 9.2724 | 12.1142 | 0.5248 | 0.6982 |
| Col4 | 9.4571 | 11.4522 | 0.5231 | 0.2508 |
| Col12 | 6.3641 | 10.5389 | 0.5897 | 0.6869 |
| Col10 | 7.9697 | 9.7949 | 0.3392 | 0.733 |
| Col9 | 7.8563 | 20.1094 | 0.6995 | 0.7432 |
| Col8 | 12.2318 | 15.4548 | 0.4203 | 0.4538 |
| Col13 | 11.0377 | 7.3343 | 0.482 | 0.4641 |
| Col15 | 15.6817 | 8.2707 | 0.8169 | 0.1343 |
| Col18 | 14.7916 | 15.5736 | 0.6674 | 0.6745 |
| Col19 | 14.2126 | 16.4898 | 0.4545 | 0.3566 |
| Col29 | 7.1853 | 16.3467 | 0.5445 | 0.5013 |
| Col26 | 9.7724 | 9.6269 | 0.7118 | 0.7865 |
| Col22 | 8.4168 | 5.5958 | 0.6049 | 0.5645 |
| Col33 | 9.1982 | 8.9987 | 0.6508 | 1.374 |
| Col34 | 10.6987 | 7.883 | 0.5067 | 0.5031 |
| Col35 | 13.8427 | 7.5469 | 0.4363 | 0.6281 |
| ConRef | 6.506 | 7.9323 | 8.7811 | 7.9323 |
Example of use
See also "Hello World project".
 "
"