|
|
| Line 73: |
Line 73: |
| | </div> | | </div> |
| | </div> | | </div> |
| - | <div class="item may">
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 4</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">The elongation of the pSB6A1-AraC-lacZ backbone turned out to be difficult. So in week 4, we performed different restriction digests both with our final backbone pSB6A1-AraC-lacZ and with the former construct pSB1C3-AraC-lacZ to verify the identity of the backbone. Besides, the amplification of the DelH fragment F1 was planned: it will be amplified in 2 subfragments - fragment F1a and fragment F1b (both 5 kb in size). </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item may last">
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 5</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">This week, we started over with the assembly of the backbone pSB6A1-AraC-lacZ: digesting AraC, lacZ and PSB6A1 based on the previously amplified fragments. The amplification of DelH was continued and we succesfully amplified all three fragments and gel-extracted them. </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item june first">
| |
| - | <img src="data:image/png;base64,"/>
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 6</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Since all DelH-fragments required for the final construct as well as the backbone were assembled in week 5, we tried to assemble the final plasmid pHM01. Therefore, every fragment was digested with two distinct enzymes, and then ligated. The ligated plasmid pHM01 was purified and electroporated in two separate DH10ß aliquots. The screening via colony-PCR was negative, so none of the transformed <i>E.coli</i> received the correct plasmid. </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item june">
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 7</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In week 7, we reamplified the DelH fragments used for the transformation in week 6. We found that amplification of fragments F1a and F1b was not reproducible. Therefor, we designed new primers for DelH, which will allow to amplify the beginning of DelH in a more efficient and specific manner. The overview summarizes the primers we ordered and their performance in amplificating DelH F1a. Primer DN11 yielded best results and will be used in the next experiments. </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item june">
| |
| - | <img src="data:image/png;base64,"/>
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 8</h1>
| |
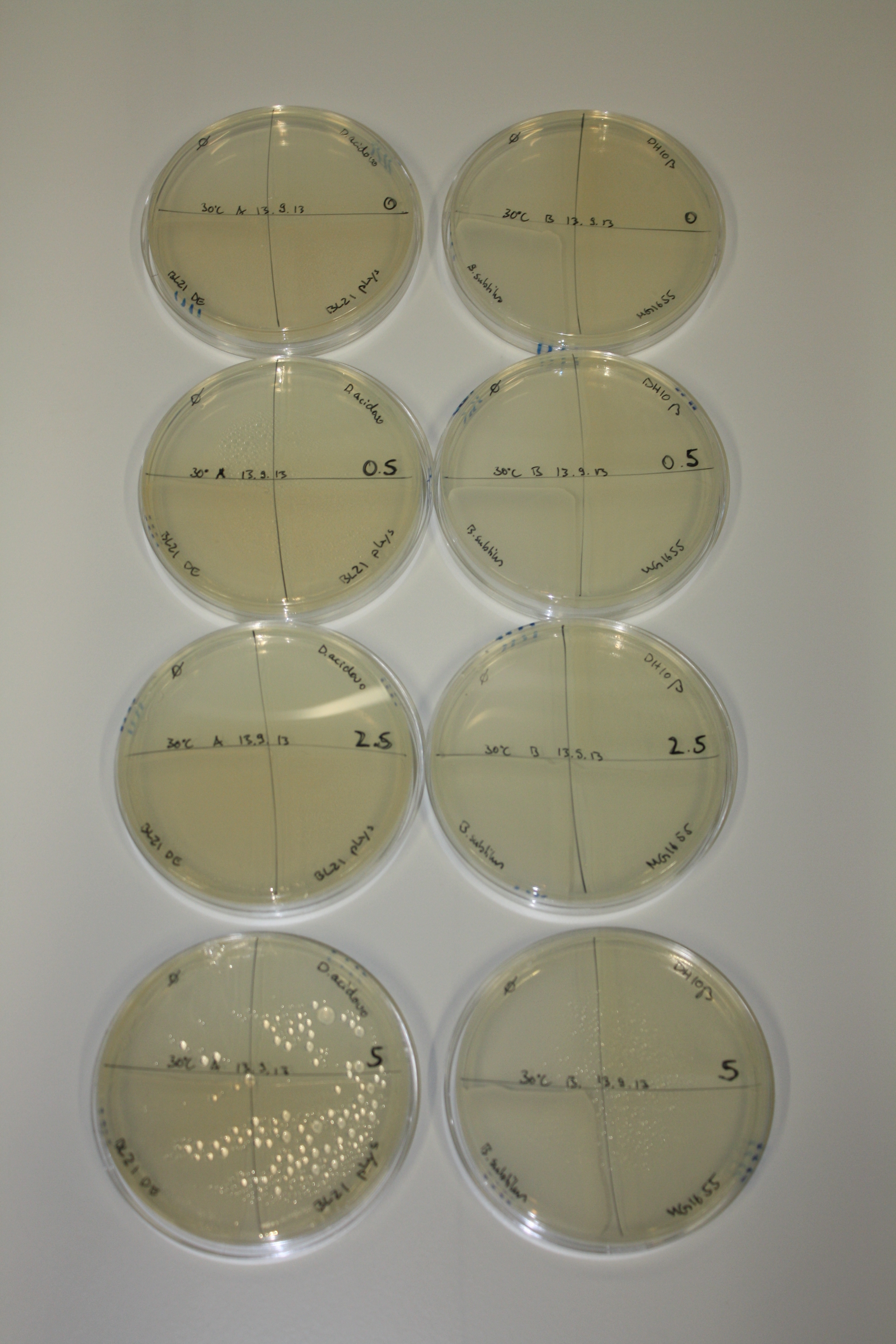
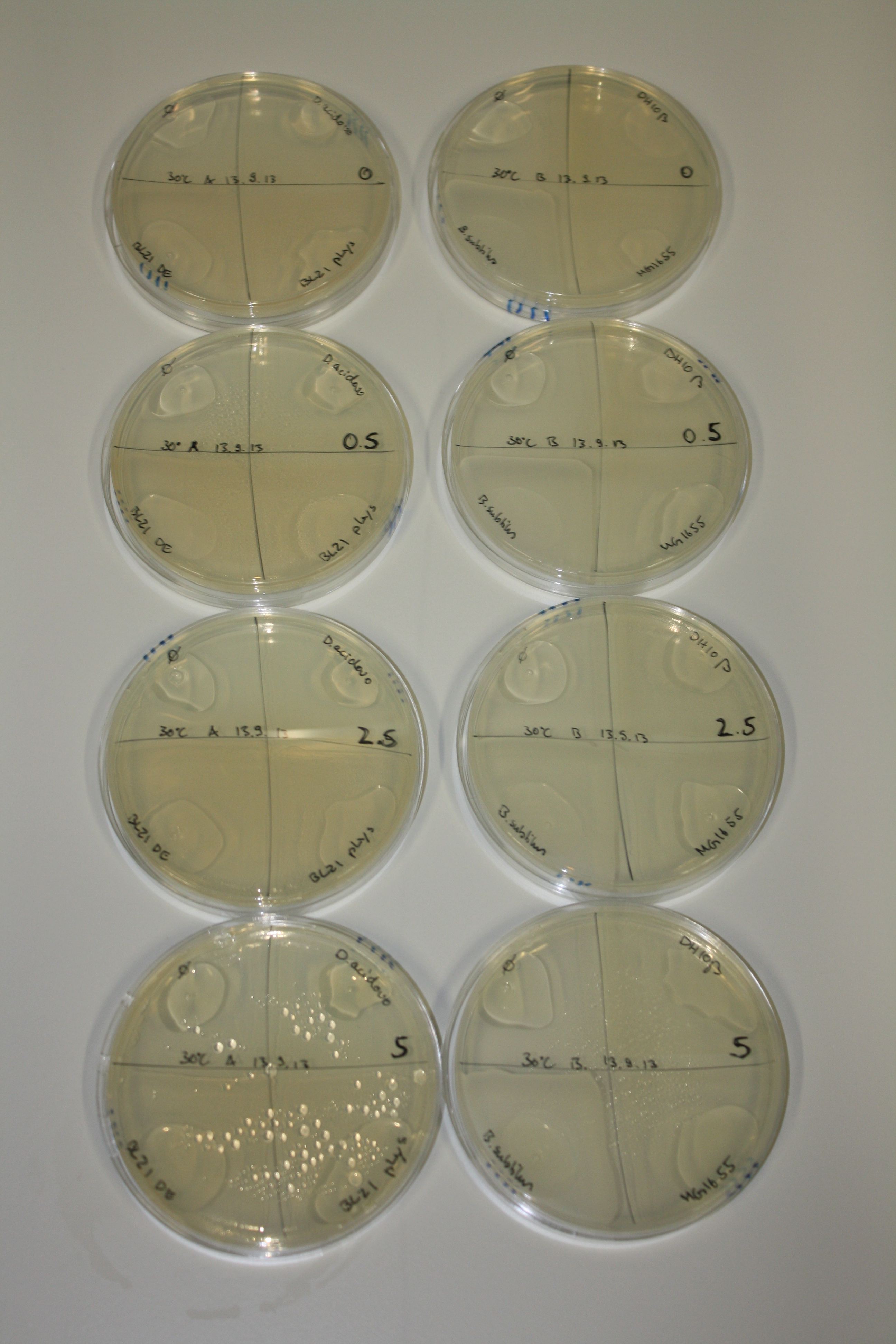
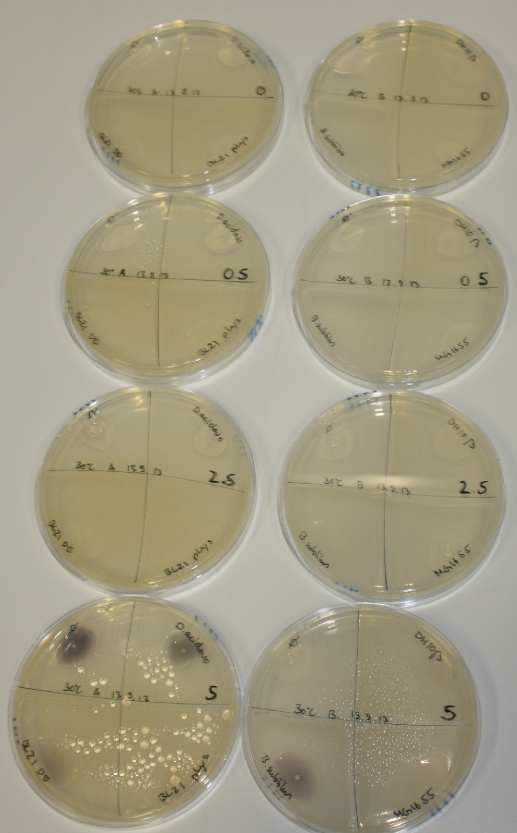
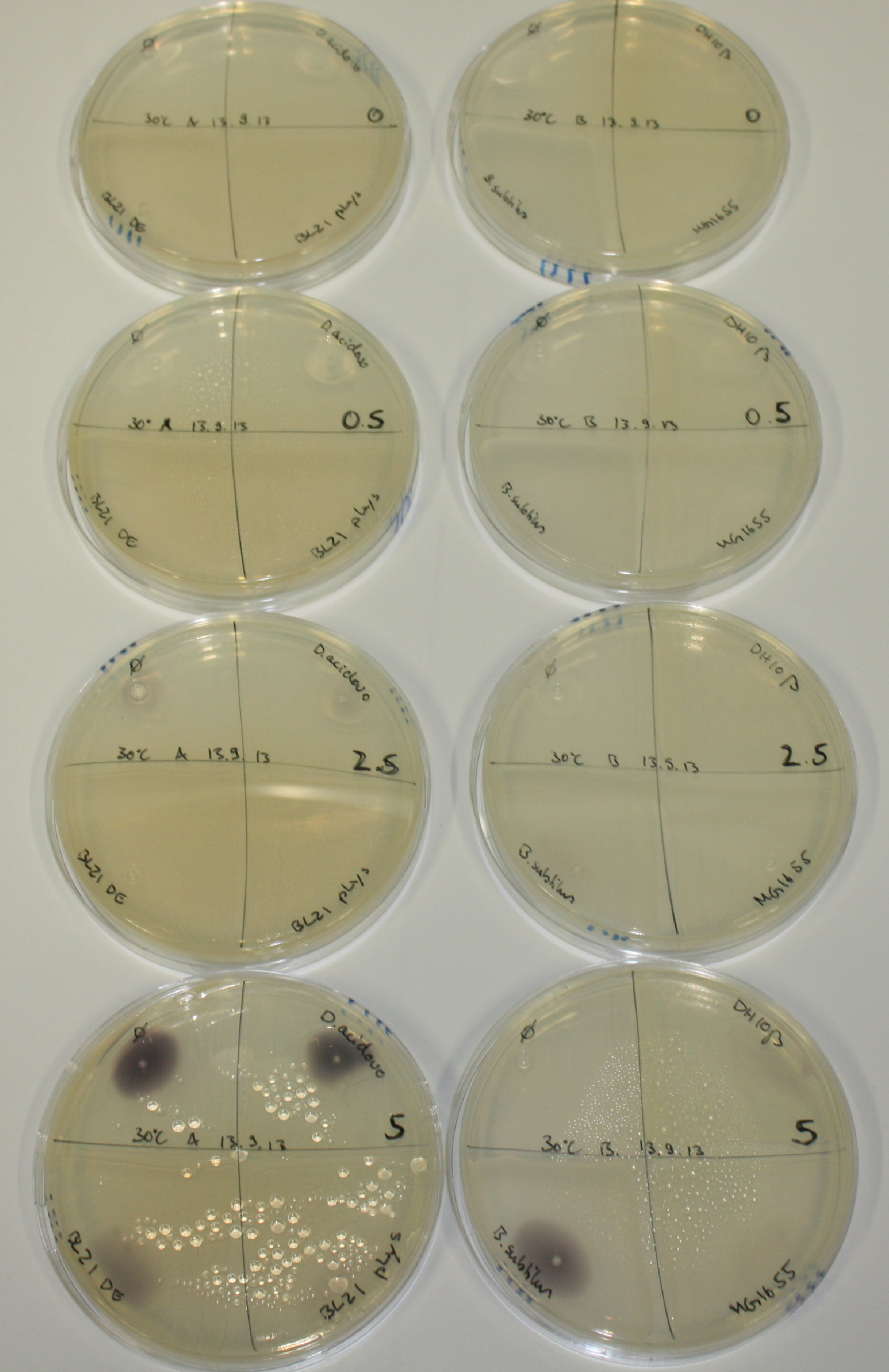
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After improving the amplification of the first subfragment of DelH (fragment 1a with the primer DN11), we used a higher concentration in the ligation assembling the pHM01 plasmid. The transformation of <i>E.coli</i> DH10ß cells was performed with electroporation. Four colonies were positive in the screening-PCR, but did not express lacZ (no blue color). To qualitatively determine DelH expression, we plan to conduct SDS-PAGEs of cell lysate derived from DelH transformed cells. Also, we will induce the transformed bacteria with higher concentrations of arabinose and X-Gal.
| |
| - | </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item june last">
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0" id="week1">
| |
| - | <h1>Week 9</h1>
| |
| | | | |
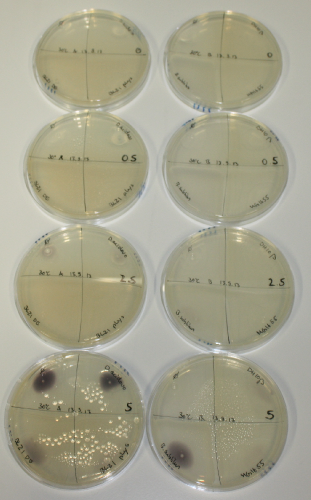
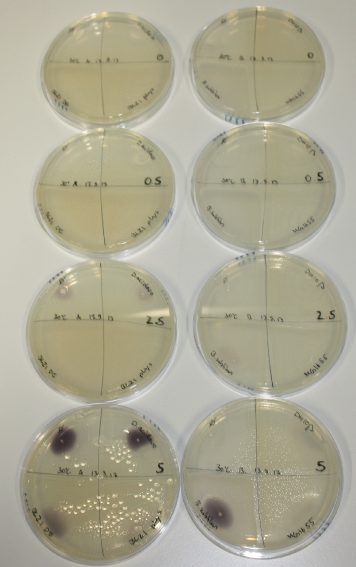
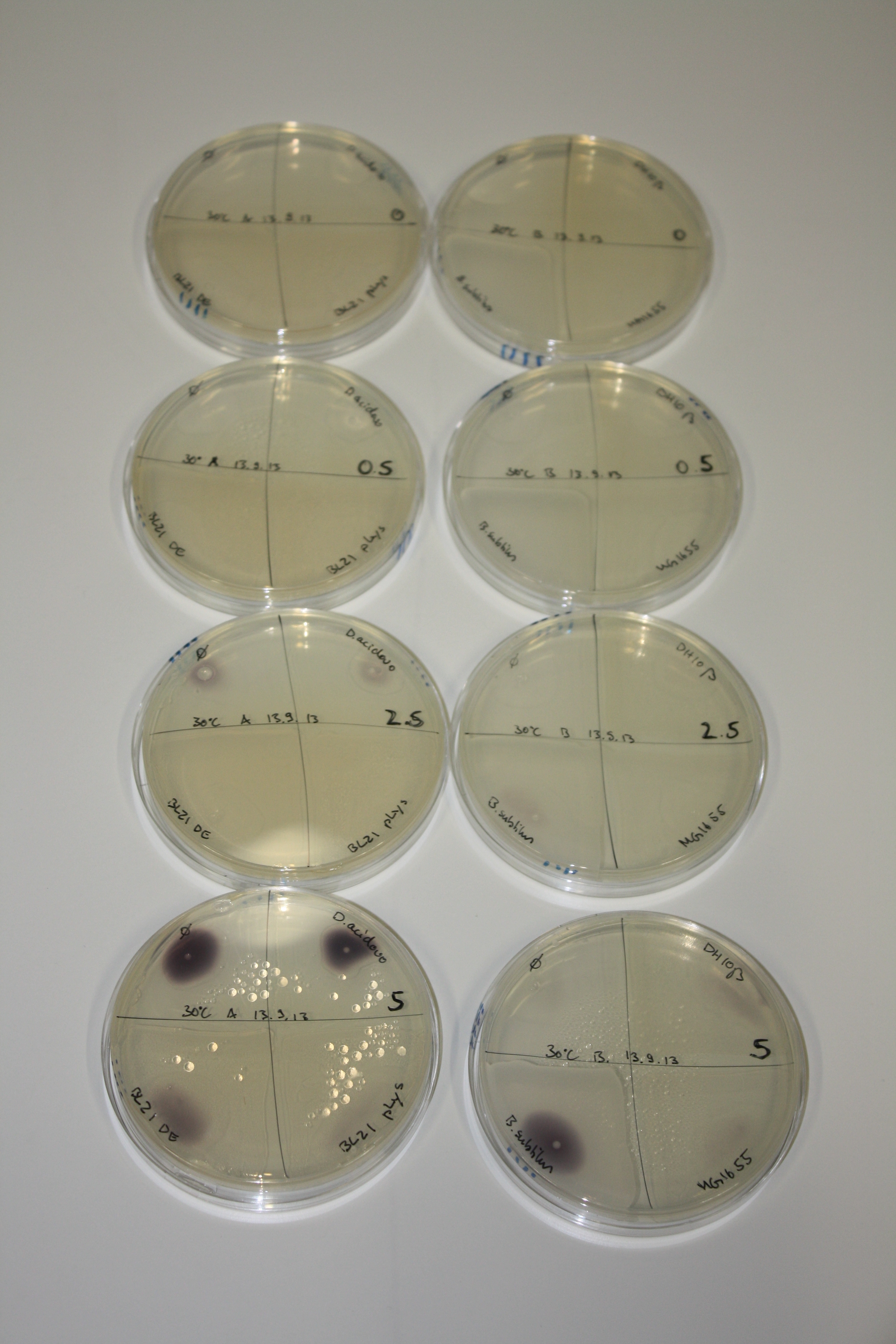
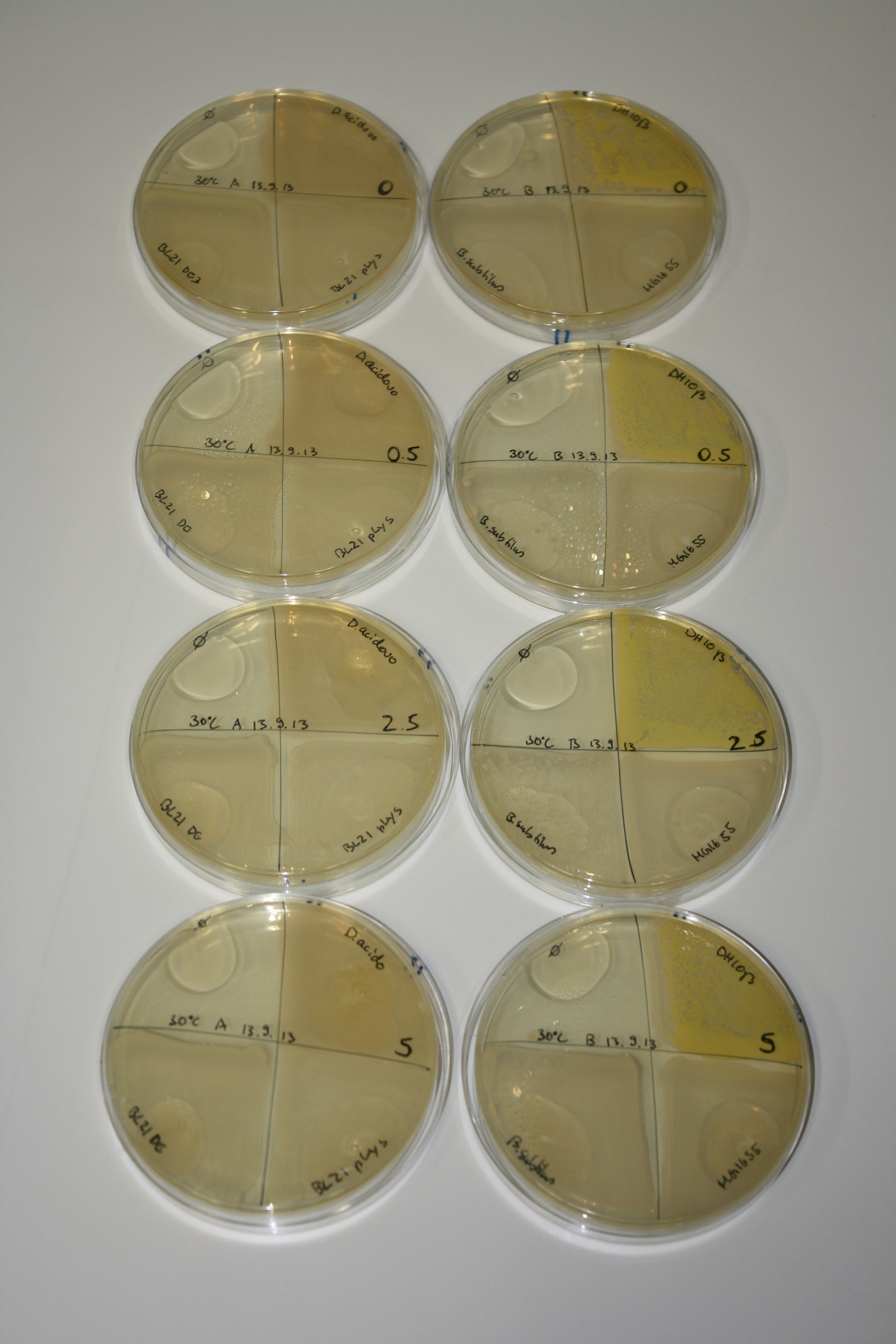
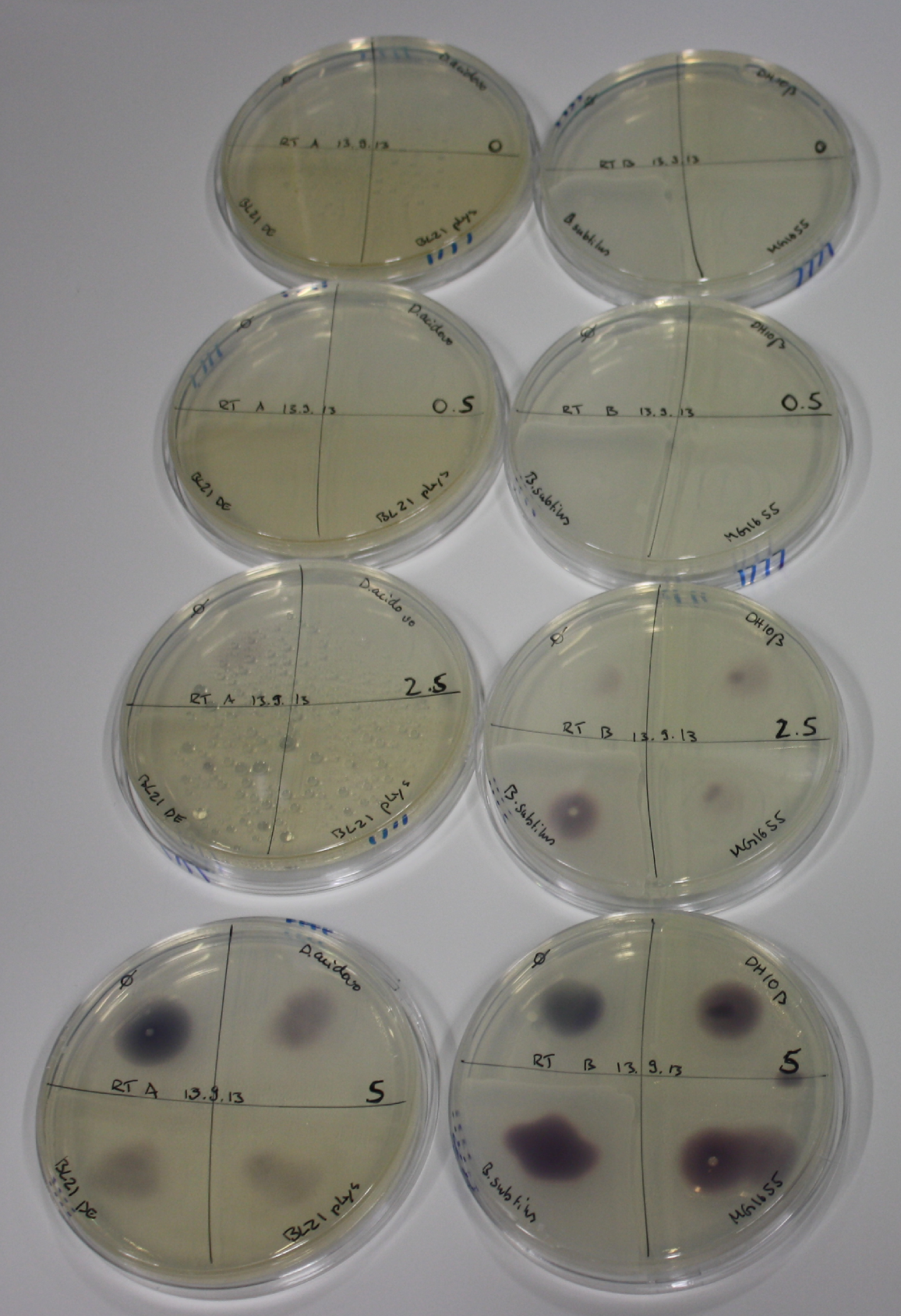
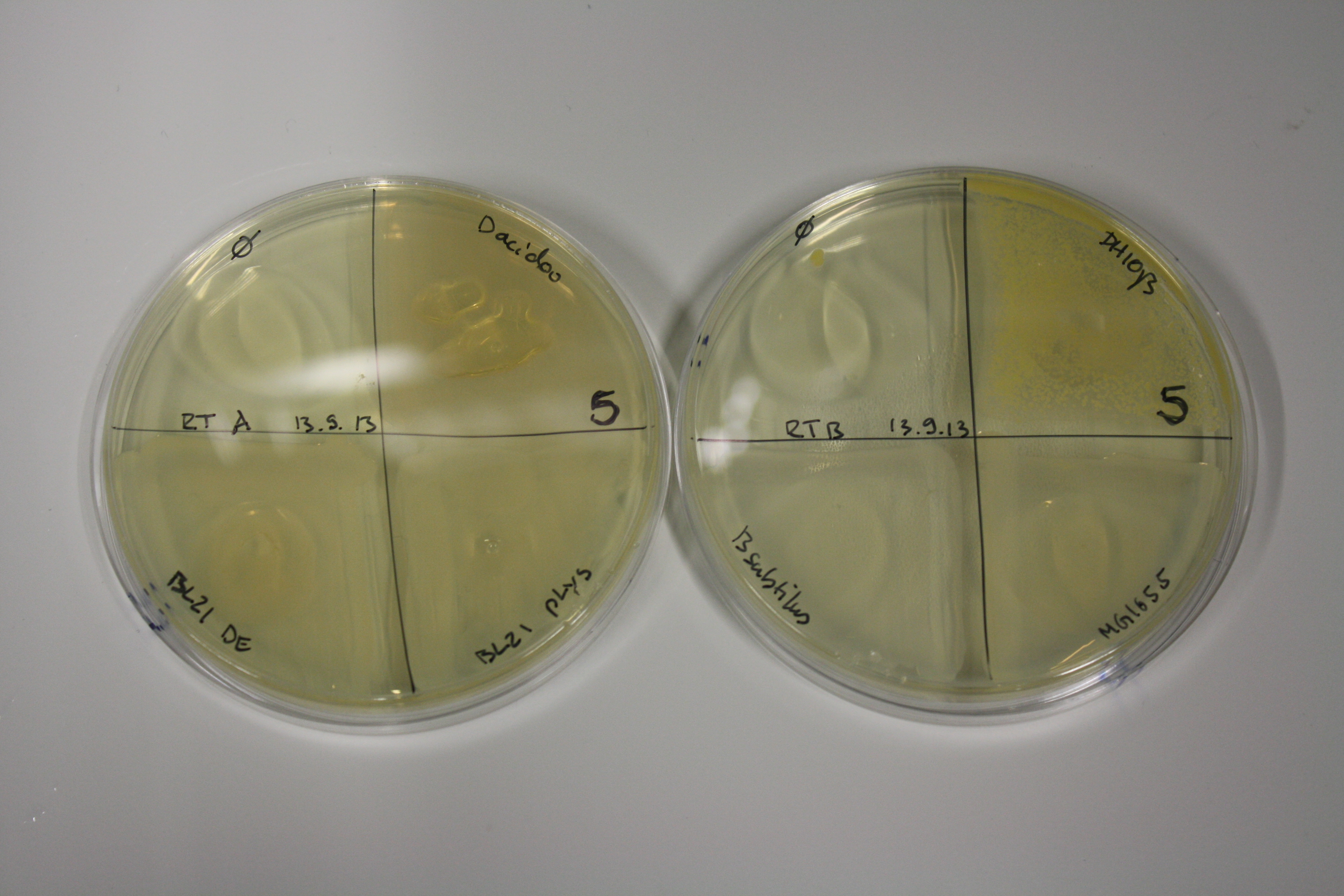
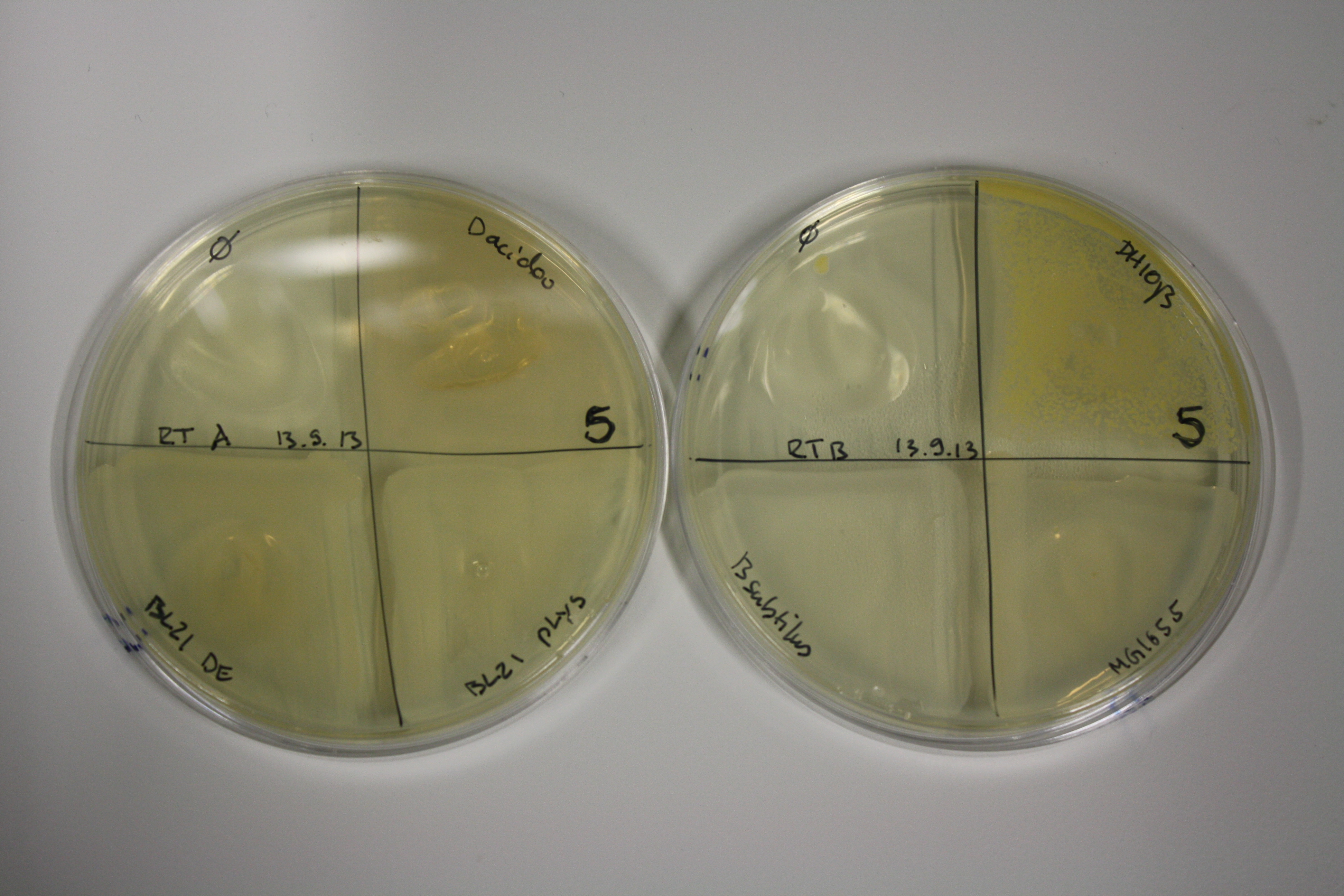
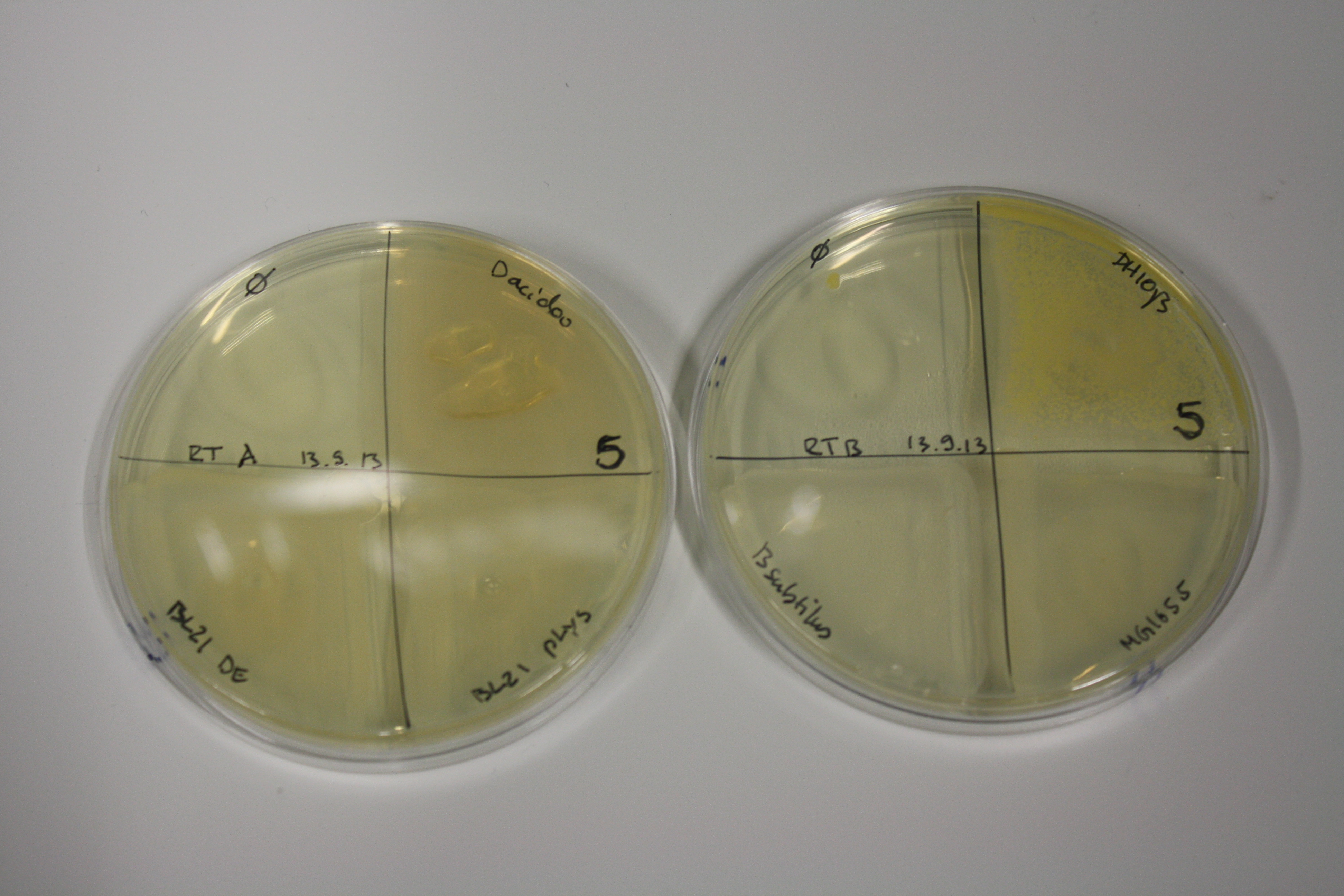
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;"> SDS-PAGE of cell lysate derived from DelH-transformed with subsequent coomassie staining did not yield a band at the expected protein size of ~600 kDa. The result was confirmed on DNA level, as the restriction digest of the DNA prepped from colonies of the transformed cells did not show the expected pattern. To perform a new assembly of plasmid pHM01, the fragments of DelH and the backbone had to be reamplified. The amplifications of the DelH fragments were successful (confirmed by gelelectrophoreses), but the backbone caused difficulties and we were not able to amplify it.
| |
| - | </p>
| |
| - |
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item july first">
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 10</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">After unsuccessful assembly attempts of pHM01, we tried to verify the previously amplified fragments. Two difficulties were encountered: On the one hand, the concentrations of the fragments were too low and could not be detected by gelelectrophoresis; on the other hand, a re-amplification of the fragments with the appropriate primers was not successful. For the DelH 1b fragment, the PCR products of the amplification from genomic DNA were slightly larger than the reamplifications.
| |
| - | </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item july">
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 11</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Based on this week's experiments and <i>in silico</i> analysis of the fragments DelH F1a, F1b and F2, we discarded the restriction digest and ligation strategy for the assembly of the DelH plasmid. Instead, we chose Gibson assembly as an alternative method, developed a strategy and designed primers accordingly.
| |
| - | </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item july">
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 12</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We started amplifying the Gibson fragments using different PCR approaches. Since we were not able to amplify G1 by PCR, we decided to divide G1 into the subfragments G1a and G1b, for which we had already designed and ordered primers. The amplification of fragment G2 worked well and yielded sufficient DNA amounts for subsequent steps. Alternatively, we also produced the entire DelH G0 as one fragment, as well as further subfragments DelH G1/2a, G1b/2 and G2b. Amplification of G1b/2a did not work out. Due to the failure of the restriction digest strategy, we further analyzed the backbone pSB6A1-AraC-lacZ, which we decided to discard. Instead, we choose to use pSB6A1 and BBa_J04450, which is already available in the parts registry. </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item july">
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 13</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In order to realize the new strategy using the already existing backbone from the parts registry pSB6A1-lacZ-mRFP, we designed two new primers for the backbone amplification and created a map of pHM03.
| |
| - | The plasmid for the backbone was obtained from the registry, transformed into <i>E.coli</i> and miniprepped. The Gibson fragment of the backbone was successfully amplified, together with Gibson fragments DelH G0, G1 and G2b. </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item july last">
| |
| - | <img src="data:image/png;base64,"/>
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 14</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">In week 14, we screened numerous colonies from last week's Gibson assembly (28-07) for plasmids containing DelH G0, G1/2a and 2b by screening-PCR. None of the analyzed colonies carried the correct plasmid. We performed another Gibson assembly (01-08) and screened numerous clones. We found few possibly correct ones that will be further analyzed next week.
| |
| - | Additionally, the new <i>D. acidovorans</i> strain SPH1, whose sequence is available in GenBank, was ordered. We will amplify all fragments from the genome of <i>D. acidovorans</i> SPH1 as soon as it arrives. </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| | <div class="item august first"> | | <div class="item august first"> |
| | <div class="container"> | | <div class="container"> |
| Line 177: |
Line 79: |
| | <h1>Week 15</h1> | | <h1>Week 15</h1> |
| | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We screened colonies from last weeks Gibson assemblies (01-08) for plasmids containing DelH G0 as well as G1/2a and 2b. None of the screening-PCRs yielded the expected DNA bands. Therefore, we amplified the Gibson fragments again and performed further Gibson assemblies. Yet again, we could not detect positive colonies by colony-PCR. </p> | | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We screened colonies from last weeks Gibson assemblies (01-08) for plasmids containing DelH G0 as well as G1/2a and 2b. None of the screening-PCRs yielded the expected DNA bands. Therefore, we amplified the Gibson fragments again and performed further Gibson assemblies. Yet again, we could not detect positive colonies by colony-PCR. </p> |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item august">
| |
| - | <img src="data:image/png;base64,"/>
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 16</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">We further characterized the DelH plasmid created by Gibson assembly. None of the screened colonies yielded definit positive results. Selection of red colonies was not clear, and PCR screened colonies were all negative.
| |
| - | In order to avoid high background during screening of the colonies, we decided to run two different strategies. First, we will amplify the backbone without mRFP, avoiding backbone reassembly due to ribosome binding site homology (pHM04). In the second strategy (pHM05), we will additionally introduce a tetracycline resistance to select for the insert via antibiotic resistance.
| |
| - | In addition to the primers for the new strategies, we designed a new screening primer at the end of DelH. </p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item august">
| |
| - | <img src="data:image/png;base64,"/>
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 17</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">A possible explanation for the failed Gibson assemblies could be a religation of the backbone fragment pSB6A1 allowing <i>E.coli</i> to survive. In this case, transformed bacteria will still express mRFP. This week, our aim is to design a new construct without mRFP (pHM04), by which we will be able to exclude red colonies from the screening. For this strategy, we are going to use a new reverse primer for the backbone still including the terminator of mRFP, but omitting mRFP itself. The primers for the backbone amplification are HM11 & HM17.</p>
| |
| - | </div>
| |
| - | </div>
| |
| - | </div>
| |
| - | <div class="item august">
| |
| - | <img src="data:image/png;base64,"/>
| |
| - | <div class="container">
| |
| - | <div class="carousel-caption scrollContent2" data-spy="scroll" data-target="#navbarExample" data-offset="0">
| |
| - | <h1>Week 18</h1>
| |
| - | <p style="font-size:10pt; text-align:justify; position:relative; margin-left:6%;">Since the assembly strategy for pHM04 did not yield positive clones (see experiments week 17), we will follow the idea to introduce a tetracycline resistance to the ampicillin backbone as an additional selection marker for successful assembly. With this approach, positive clones can be easily determined by their white phenotype (exclusion of mRFP) and their ability to grow on plates containing tetracycline. Furthermore, we decided to amplify DelH in various fragments to increase Gibson assembly efficiency. Therefore, we also ordered new screening primers. The primers were designed as shown in the following table. </p>
| |
| | </div> | | </div> |
| | </div> | | </div> |


 "
"