Team:Valencia Biocampus/Project
From 2013.igem.org
Beta3designs (Talk | contribs) (→Results) |
|||
| Line 179: | Line 179: | ||
<div class="clearfix"></div> | <div class="clearfix"></div> | ||
<p> | <p> | ||
| - | <h2>Why use C. elegans as a transport?</h2> | + | <h2>Why use <i>C. elegans</i> as a transport?</h2> |
</p> | </p> | ||
<p> | <p> | ||
| Line 206: | Line 206: | ||
</p> | </p> | ||
<p> | <p> | ||
| - | <h2>Expanding our knowledge about C. elegans...</h2> | + | <h2>Expanding our knowledge about <i>C. elegans</i>...</h2> |
</p> | </p> | ||
<p> | <p> | ||
| Line 244: | Line 244: | ||
<p> | <p> | ||
| - | + | In this synthetic symbiosis<b>, <i>C. elegans</i> </b>acts as a transport for engineered bacteria <b>(<i>Pseudomonas putida</i></b><i>) </i>in order to | |
| - | + | take them to the hotspot of interest, because bacteria are are not able to move fast in through solid or semi-solid substrates but they are very | |
| - | + | interesting from a biotechnological point of view, that is the reason why we though in this innovative mean of transport: the regulated <b> formation of a biofilm</b>. | |
</p> | </p> | ||
<p> | <p> | ||
| - | To achieve our goal, we constructed a BioBrick (see part: <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1112001">BBa_K1112001</a>) | + | To achieve our goal, we constructed a BioBrick (see part: <u><a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1112001">BBa_K1112001</a></u>) |
| - | consisting in the coding sequence of the hmsHFRS operon, an adhesion operon natural from <i>Xenorhadbus nematophila</i> which allows the formation of a | + | consisting in the coding sequence of the <b>hmsHFRS operon</b>, an adhesion operon natural from <i>Xenorhadbus nematophila</i> which allows the formation |
| - | + | of a biofilm on the nematode <i>S. carpocapsae</i>, under the control of a nitrogen sensitive promoter (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K763003"><b>pGlnA, </b>characterized by the <b> 2012 Valencia Biocampus iGEM team</b></a>). (<b>Fig.1</b>) | |
| - | + | ||
</p> | </p> | ||
| - | <p> | + | <p style="text-align:center"> |
| - | + | <img src="https://static.igem.org/mediawiki/2013/1/1b/Vb_riding_1.png" style="width:700px;" alt="" /> | |
| - | + | ||
| - | + | ||
| - | + | ||
</p> | </p> | ||
<p> | <p> | ||
| - | + | <h3>Controlling the mechanism</h3> | |
| - | + | Riding was a regulated process: with <b>low nitrogen </b>in the media, the promoter is activated and <i>hms</i> genes are expressed, triggering the <b>formation of the biofilm </b>over <i>C. elegans</i>; in contrast, with <b>high nitrogen </b>concentrations, such as the ones found in nutrient-rich hotspots, the promoter is repressed, so bacteria can <b>“get off</b>” the nematode <b>(Fig. 2)</b>. | |
| - | + | ||
</p> | </p> | ||
<p style="text-align:center"> | <p style="text-align:center"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | <img src="https://static.igem.org/mediawiki/2013/2/20/Vb_riding_2.png" style="width:700px;" alt="" /> |
</p> | </p> | ||
<p> | <p> | ||
| - | + | <h3>Biofilm formation in genetically-engineered bacteria</h3> | |
| + | |||
| + | Our original idea was to introduce the Biobrick in <b><i>Pseudomonas putida </i></b>, a bacterial species with wide applications in biotechnology. To do | ||
| + | that, we cloned the construction (<b>Fig.1)</b> in the <b>pIZ1016 vector</b>, which has a replication origin compatible with <i>Pseudomonas</i>. We | ||
| + | successfully performed the cloning <b>(Fig.3)</b>, but the efficiency of the transformation was too low, so haven’t been able to obtain <i>P. putida </i> | ||
| + | transformants yet. This is probably a consequence of the length of the construction, 6,5 kb, which decreases transformation efficiency. | ||
| + | </p> | ||
| + | <p style="text-align:center"> | ||
| + | <img src="https://2013.igem.org/File:Vb_riding_3.png" style="width:700px;" alt="" /> | ||
</p> | </p> | ||
<p> | <p> | ||
| - | + | But far from being disheartened, we decided to express the construction with <b><i>E. coli.</i> </b>We cloned the construction in the pUC57 vector, | |
| + | obtained transformant <i>E. coli</i>, and then grew them in medium with low nitrogen (0,6 g/L) in order to induce the formation of the biofilm.<i>C. elegans </i>was fed with these induced bacteria, and then several worms were isolated in order to check biofilm formation with <b>scanning electron microscopy (SEM) </b>imaging. As you can see in <b>Fig.4</b>, we actually did it! We observed a formation of an <i>E. coli </i>biofilm | ||
| + | over the nematode! | ||
</p> | </p> | ||
<p style="text-align:center"> | <p style="text-align:center"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | <img src="https://static.igem.org/mediawiki/2013/0/03/Vb_riding_4.png" style="width:700px;" alt="" /> |
</p> | </p> | ||
| - | |||
</div> | </div> | ||
<div> | <div> | ||
| Line 284: | Line 288: | ||
<p> | <p> | ||
<h3>Looking for an attractant</h3> | <h3>Looking for an attractant</h3> | ||
| - | When we were considering setting up a transport of bacteria was thought to be necessary to have a 'destination', a place to go. That | + | When we were considering setting up a transport of bacteria was thought to be necessary to have a 'destination', a place to go. That destiny would be a <b>'hot spot' on a heterogeneous substrate</b> where <b><i>Caenorhabditis</i> <i>elegans</i></b> should lead right to that point the bacteria. |
<br/> | <br/> | ||
| - | Leveraging the powerful smell of the nematode, it was decided to try a number of attractants from various lists | + | Leveraging the powerful smell of the nematode, it was decided to try a number of attractants from various lists from web <a href="http://www.wormbook.org">www.wormbook.org</a> that could work as 'hot spot' of our experiment. Thus, using the<i> C. elegans</i> <b>chemotaxis, </b>we could direct transport. |
</p> | </p> | ||
| Line 293: | Line 297: | ||
<h3>The attractants experiment</h3> | <h3>The attractants experiment</h3> | ||
| - | The test would be carried creating our own plates on which | + | The test would be carried creating our own plates on which half would be NGM unmodified and the other half part would be including soluble compounds before |
| - | solidifying or after solidification | + | solidifying or after solidification in the case of volatiles. The list of modifications can be found in <b>Fig. 1</b>. |
</p> | </p> | ||
<p style="text-align:center"> | <p style="text-align:center"> | ||
| Line 300: | Line 304: | ||
</p> | </p> | ||
<p> | <p> | ||
| - | To place <b><i>C. elegans</i> </b>on the plate, different cuts were made on a fresh plate of NGM | + | To place <b><i>C. elegans</i> </b>on the plate, different cuts were made on a fresh plate of NGM, the resulting small pieces were placed in the exact |
| - | center of the 50% -50% plates to determine which side of the | + | center of the 50% -50% plates to determine which side of the nematode preferred, one per Petri plate. |
</p> | </p> | ||
<p> | <p> | ||
| Line 310: | Line 314: | ||
</p> | </p> | ||
<p> | <p> | ||
| - | From <b>Table 2 </b> | + | From <b>Table 2 </b>it could rule out many of the attractants that were thought viable. |
</p> | </p> | ||
<p> | <p> | ||
| - | Volatile attractants were not a good choice | + | Volatile attractants were not a good choice to evaporate quickly (which is also limited to the field experiments). |
</p> | </p> | ||
<p> | <p> | ||
| - | Another impediment arose. Once | + | Another impediment arose. Once had already performed the experiments, it was decided that the promoter that will control the production of RNA interference |
| - | in <i>Escherichia coli</i> would be controlled by nitrogen, amino acids had to be | + | in <i>Escherichia coli</i> would be controlled by nitrogen, amino acids had to be discard as attractants; it would modify controlled expression of E.coli. |
</p> | </p> | ||
<p> | <p> | ||
| Line 323: | Line 327: | ||
</p> | </p> | ||
<p> | <p> | ||
| - | <h3> | + | <h3>Attractant final choice</h3> |
| - | Because the hyposmotic medium could interfere with the proper growth of bacteria (food | + | Because the hyposmotic medium could interfere with the proper growth of bacteria (food of our nematode), the final decision was to choose the MgSO<sub>4</sub> as attractant. |
</p> | </p> | ||
<p> | <p> | ||
| - | The question that arose at that time was: <i>How | + | The question that arose at that time was: <i>How we can multiply the amount of MgSO<sub>4</sub> to increase efficiency?</i> |
</p> | </p> | ||
<p> | <p> | ||
| Line 358: | Line 362: | ||
<p> | <p> | ||
<h3>Final concentration choice</h3> | <h3>Final concentration choice</h3> | ||
| - | Once the test | + | Once the test already performed and that the results seen by the factor of PHA and MgSO<sub>4</sub> selected fatty acid give very scattered (there is |
| - | probably repellent effect at high concentrations in the medium with oleic but reversed in octanoic) we | + | probably repellent effect at high concentrations in the medium with oleic but reversed in octanoic) we make a selection of MgSO<sub>4</sub> concentration |
for each medium. | for each medium. | ||
<ul> | <ul> | ||
| Line 498: | Line 502: | ||
detection (complexes of PHA with red Nile are fluorescent), its bright future in the field of biomaterials and because we were using this natural ability | detection (complexes of PHA with red Nile are fluorescent), its bright future in the field of biomaterials and because we were using this natural ability | ||
of bacteria as a tool. | of bacteria as a tool. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | <h3>The mechanism</h3> | ||
| + | |||
| + | A cluster of genes is responsible of the production of PHA, the <b>phaC operon </b>of <b><i>P. putida </i></b>which consists of 4 ORFs that are transcribed | ||
| + | in the same direction: phaC1 and phaC2 genes, codify for two PHA sintases; phaZ gene, codifies for a despolimerase; and phaD gene, that codifies for a | ||
| + | protein of the TetR family. In the opposite direction two genes that codify for fasinas (phaF and phaI) and structural proteins are found <b>(Fig.1). </b> | ||
</p> | </p> | ||
Revision as of 14:33, 4 October 2013
Project Overview
Why E.coli
Escherichia coli is a model organism widely use in fields as microbiology, molecular biology and genetics. Result of that is the great range of genetic manipulation techniques that can be found related to this Gram negative bacterium. There is also a lot of information about the biochemistry and genetics of this microorganism. All this allows to work easily with it in the lab and often perform assays testing different conditions that will be tried out in other organisms unstudied in such depth. Moreover, E.coli is the main food of C.elegans, organism that is able to incorporate nucleic acids from the bacteria, RNA, to its cells (Mello et al., 2004). This is the usual mechanism of transformation of the worm, introducing exogenous genetic material from the bacteria altering the expression of the worm’s genome in order to make modifications of interest.
Controlling the mechanism
The role of E.coli in our project is to carry out the synthesis of an iRNA for inducing the social feeding behavior of Caenorhabditis elegans , the clumping. To achieve this, we cloned the biobrick Bba_K1112000 in E. coli, XL1-Blue strain (fig. 1).

Fig. 1.
- pSB1C3 is a high copy number plasmid (RFC [10]) carrying chloramphenicol resistance.
- The replication origin is a pUC19-derived pMB1 (copy number of 100-300 per cell).
- pSB1C3 has terminators bracketing its MCS which are designed to prevent transcription from *inside* the MCS from reading out into the vector. The efficiency of these terminators is known to be < 100%. Ideally we would construct a future set of terminators for bracketing a MCS that were 100% efficient in terminating both into and out of the MCS region.
This construction consists of the E.coli fadA promoter, which is activated in the presence of fatty acids (Clark, 1981), and the antisense sequence of the mRNA FLP-21 from C.elegans which encodes a protein involved in the solitary feeding behaviour of the worm, the formation of the dsRNA complex inhibits the expression of this protein, inducing the social feeding behavior (fig. 2).

Pseudomonas putida and PHA production
Pseudomonas putida is a gram-negative bacterium that is found in most soil and water habitats where there is oxygen. Its diverse metabolism and its capacity to break down organic harmful solvents (it has most genes involved in degrading aromatic or aliphatic hydrocarbons) in contaminated soils make this microorganism irreplaceable for research studies in the field of bioremediation but also for biosynthesis of value-added products. In addition, Pseudomonas putida has several strains including KT2440, the one we have worked with. This strain can colonize plant roots, from which they take nutrients, while at the same time it offers protection to the plant from pathogens.
For example, it is capable of converting styrene oil into the biodegradable plastic PHA. This helps to degrade the polystyrene foam which
was thought to be non-biodegradable. Styrene is a major environment toxic pollutant released from industrial sites. The conversion to PHA allows the cure
of styrene pollution but it is also beneficial to society because of its applications in tissue engineering.
PHA is also environmental friendly and has a long self-life therefore it is also used in everyday items. Unlike styrene, PHA can break down in soil or water.
Within Pseudomonas putida, PHA accumulates under unbalanced growth conditions as a means of intracellular storage, storing excess carbon and energy. These PHA polymers are synthesized by enzyme PHA synthase which is bound to the surface of the PHA granules and uses coenzyme A thioesters of hydroxyalkanoic acids as substrates.

The role of P. putida in the Synthetic symbiosis that we have designed is to be carried by C. elegans to hotspots of interest where would produce bioplastic PHA.
An overview about our nematode
One of the main characters of our work is a nematode known as Caenorhabditis elegans, from the Rhabditidae family. It was first used as an experimental model in Developmental Genetics studies and nowadays is also used in other fields such as Clinical Biology, Neurobiology and Cell Biology, being a good model to study Alzheimer disease, obesity, diabetes and aging, among others.
Another interesting thing is that it feeds on Escherichia coli. Its “favorite” strain is OP50, although we checked that it’s also able to feed on XL1-Blue strain, the one that we used in all our molecular biology experiments.
Some advantages of C. elegans when compared with other model organisms are:
- Lifespan ranges between 2 and 3 weeks, so experimentation times are reduced.
- Its maintenance and study is cheap and simple (it is transparent, which facilitates microscopic observation).
- It is very small (1 mm), so it is possible to carry out experiments with a huge number of worms in a small Petri dish having a great statistical support.
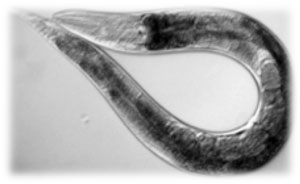
Why use C. elegans as a transport?
When we were considering on creating a new system for the transport of bacteria, we found different key advantages that made the nematode the best option. For example, C. elegans is able to move very fast around solid substrates (soil is, actually, its natural habitat) and also in agar, so it’s very useful for both lab experiments and real-environment tests.
Its movement, in addition to being very fast, has two modes: random and directed.
When there is no attractant in the medium, C. elegans moves doing uncoordinated movements in several directions in what is known as 'random walk'.
The situation changes when there is an attractant in the medium. Here, our 'transport' begins to direct his movements to the focus of the substance (volatile or soluble) which acts as an attractant in a process known as 'chemotaxis' thanks to the amazing smell of our nematode. This allows us to ‘guide’ the nematodes towards defined spots in irregular substrates.
Chemotaxis is the foundation to guide the transport of bacteria and is therefore the focus of experimentation with C. elegans, with the aim of finding the best attractant. This ability makes C. elegans a perfect 'bus' for bacteria. (simuelegans online here)
Moreover, we found two pathogens (Yersinia pestis and Xhenorhabus nematophila) with the ability to form biofilms on C. elegans thanks to the proteins of the operon hmsHFRS. When genetically-engineered strains of commonly used bacteria such as Escherichia coli or Pseudomonas putida express this hmsHFRS operon, they have the 'ticket' to travel: they are able to adhere to the worm’s surface by means of forming a synthetic biofilm.
Expanding our knowledge about C. elegans...
Once we knew that C. elegans was the best option, we began to discover interesting things for further study.
There are several strains of our nematode. The one that drew our attention was called 'N2', which had a fairly interesting behavior: under normal conditions, it eats individually, whereas under certain conditions (such as starvation), a social feeding behavior known as “clumping” is induced. But this phenomenon can also be induced if the expression of some particular genes is interfered. This fact gave us the opportunity to develop the first artificial symbiosis between worms and bacteria, based on the manipulation of the behaviour of C. elegans by simply nourishing it with transformed E.coli able to synthesize the iRNA.
With it, while C. elegans acts as transport, bacteria return the favour giving it the ability to eat in company.

Results
Parts
These are the BioBricks we have designed, constructed, and characterized. We have submitted them to the Registry of Standard Biological Parts
| Works? | Name | Type | Description | Designer | Length | Structure |
|---|---|---|---|---|---|---|
| BBa_K1112000 | Regulatory | fadB promoter + FLP-21 iRNA | Pedro Luis Dorado Morales | |||
| BBa_K1112001 | Regulatory | pGlnA + hmsHFRS operon | Alba Iglesias Vilches | |||
| BBa_K1112002 | DNA | Cluster PHA | Alba Iglesias Vilches |
BBa_K1112000: fadB promoter + FLP-21 iRNA
This construction is made up by a fatty acid-sensitive promoter that directs the transcription of an iRNA responsible for the social or solitary behavior of C.elegans.
BBa_K1112001: pGlnA + hmsHFRS operon
We reused our nitrogen sensitive promoter from one of our last year constructs and we employ it to control the expression on the operon that triggers the formation of a biofilm over C.elegans.
BBa_K1112002: cluster PHA
This BioBrick contains the complete natural sequence that induces the production of PHA in Pseudomonas putida (KT2440).
 "
"


























