Team:DTU-Denmark/Notebook/30 June 2013
From 2013.igem.org
(→208 lab) |
|||
| Line 1: | Line 1: | ||
| + | {{:Team:DTU-Denmark/Templates/StartPage|30 June 2013}} | ||
=208 lab= | =208 lab= | ||
| - | + | <hr/> | |
== Main purposes today == | == Main purposes today == | ||
| + | <hr/> | ||
New PCR with x7-polymerase. | New PCR with x7-polymerase. | ||
| - | |||
==who were in the lab== | ==who were in the lab== | ||
| + | <hr/> | ||
Kristian | Kristian | ||
==Procedure== | ==Procedure== | ||
| + | <hr/> | ||
Made PCRs on pZA21, GFP SF TAT, GFP SF Sec and RFP all samples where made in duplicates. | Made PCRs on pZA21, GFP SF TAT, GFP SF Sec and RFP all samples where made in duplicates. | ||
| Line 21: | Line 24: | ||
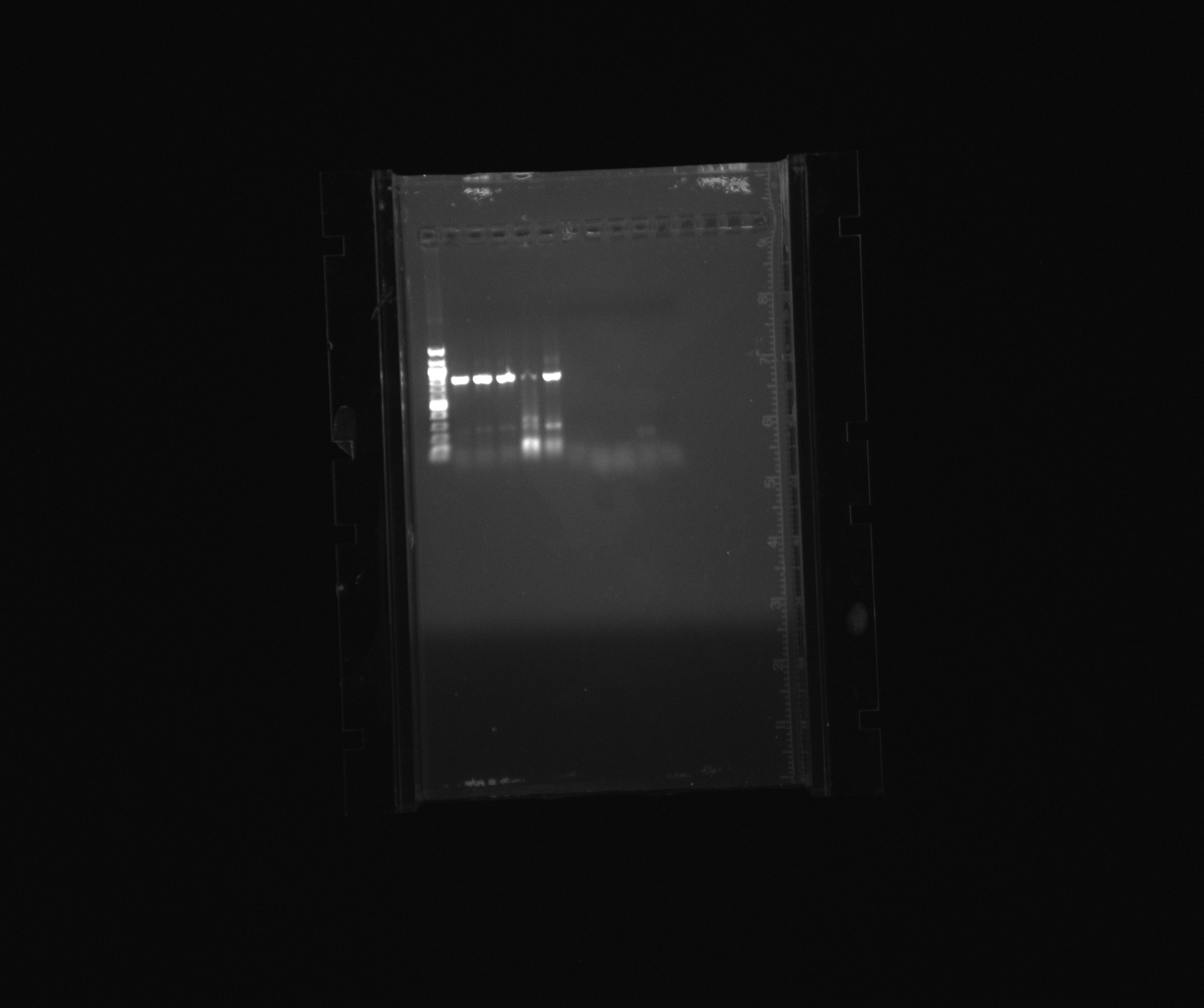
Purification was done by gel purification, see picture below. | Purification was done by gel purification, see picture below. | ||
| - | [[File:30.06.13 all PCR products purification gel.jpg|thumb|left|Purification gel with big well number one(left uppermost) containing 1+2 next 3+4, 5+6, 7+8, 9+10, 11+12, next line from left to right; 13+17, 14+16, failed well, 21+22, 24]] | + | [[File:30.06.13 all PCR products purification gel.jpg|thumb|250px|left|Purification gel with big well number one(left uppermost) containing 1+2 next 3+4, 5+6, 7+8, 9+10, 11+12, next line from left to right; 13+17, 14+16, failed well, 21+22, 24]] |
| - | + | <html><p><br/> </p></html> | |
After all PCR-products were purified the backbone was prepared for DpnI treatment and the right amount of each of the other PCR-products were mixed. Finally the DpnI treated backbone was added USER-enzyme and BSA, mixed with the appropriate PCR-products and set for reaction. | After all PCR-products were purified the backbone was prepared for DpnI treatment and the right amount of each of the other PCR-products were mixed. Finally the DpnI treated backbone was added USER-enzyme and BSA, mixed with the appropriate PCR-products and set for reaction. | ||
The constructs were transformed into chemical competent E.coli right after construction and incubated 2 hours in SOC before plated on Kana plates. | The constructs were transformed into chemical competent E.coli right after construction and incubated 2 hours in SOC before plated on Kana plates. | ||
| - | + | <html><p><br/><br/><br/><br/> </p></html> | |
==Results== | ==Results== | ||
| - | + | <hr/> | |
| - | [[File:30.06.13 all PCR products with x7 first part.jpg|thumb|left|The gel from the days PCR. From first lane is a 100 bp ladder and thereafter the well number follows the numbers of the tubes -1. Note that tube number 15 is missing]] | + | [[File:30.06.13 all PCR products with x7 first part.jpg|thumb|250px|left|The gel from the days PCR. From first lane is a 100 bp ladder and thereafter the well number follows the numbers of the tubes -1. Note that tube number 15 is missing]] |
| - | [[File:30.06.13 all PCR products with x7 second part.jpg|thumb|left|The gel from the days PCR. From first lane is a 100 bp ladder and thereafter tube 20,21,22,23 and 24. Note that there is also primer blur in subsequent wells to the right; these are just double test wells to test previous PCR products]] | + | [[File:30.06.13 all PCR products with x7 second part.jpg|thumb|250px|left|The gel from the days PCR. From first lane is a 100 bp ladder and thereafter tube 20,21,22,23 and 24. Note that there is also primer blur in subsequent wells to the right; these are just double test wells to test previous PCR products]] |
| - | + | <html><p><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> </p></html> | |
== Conclusion from today == | == Conclusion from today == | ||
| + | <hr/> | ||
All PCR-products have been made and the programs for each product have been determined. | All PCR-products have been made and the programs for each product have been determined. | ||
| + | |||
| + | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Revision as of 08:15, 12 July 2013
30 June 2013
Contents |
208 lab
Main purposes today
New PCR with x7-polymerase.
who were in the lab
Kristian
Procedure
Made PCRs on pZA21, GFP SF TAT, GFP SF Sec and RFP all samples where made in duplicates.
In tube 1+2+3+4+5+6 where pZA21. In 7+8+9+10+11+12 GFP SF TAT In 13+14+15+16+17+18 GFP SF Sec In 19+20+21+22+23+24 RFP
First program was 45°C annealing and 2:00 extension time with tube: 1+2, 7+8, 13+14, 19+20. Second program was ramp 68°C → 60°C annealing and 2:00 extension time with tube: 3+4, 9+10, 15+16, 21+22. Last program was a touch up program with 60°C annealing and 10 cycles with a temperature increment of 0.5°C per cycle this was looped 5 times so the final amount of annealing cycles was 50. Extension time was still 2:00 and this program was done on tube: 5+6, 11+12, 17+18, 23+24.
Purification was done by gel purification, see picture below.
The constructs were transformed into chemical competent E.coli right after construction and incubated 2 hours in SOC before plated on Kana plates.
Results
Conclusion from today
All PCR-products have been made and the programs for each product have been determined.
 "
"