Team:Heidelberg/Templates/Indigoidine week12
From 2013.igem.org
(Created page with " ===Indigoidine production pKH and pMM (Konrad)=== ==== IPTG Induction ==== [[File:080713-IPTG-induction.png|500px|thumb|right|Induction of 2 days old cultures with/-out 1 mM IP...") |
|||
| Line 3: | Line 3: | ||
===Indigoidine production pKH and pMM (Konrad)=== | ===Indigoidine production pKH and pMM (Konrad)=== | ||
==== IPTG Induction ==== | ==== IPTG Induction ==== | ||
| - | [[File: | + | [[File:Heidelberg_080713-IPTG-induction.png|500px|thumb|right|Induction of 2 days old cultures with/-out 1 mM IPTG. Rosetta |
seems to produce indigoidine, even without induction (leaky exppression?).]] | seems to produce indigoidine, even without induction (leaky exppression?).]] | ||
| Line 23: | Line 23: | ||
enrichment of desired fusion product with fw primer of fragment 1 and rv primer of fragment 2. | enrichment of desired fusion product with fw primer of fragment 1 and rv primer of fragment 2. | ||
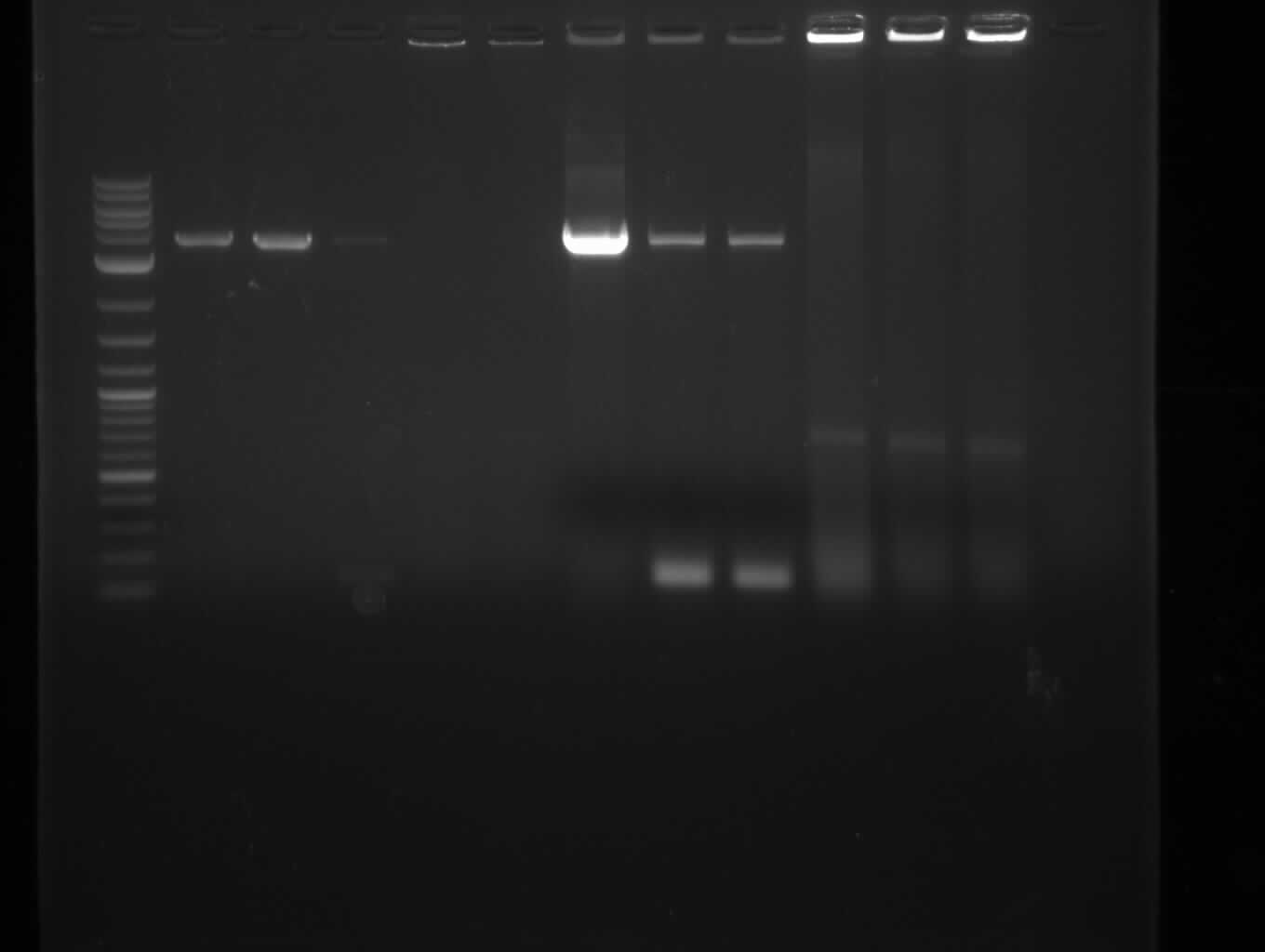
| - | [[File: | + | [[File:Heidelberg_090713-pca.png|200px|thumb|right|PCA in order to gain bpsA and svp as one fragment fused together. The |
wanted fragment size is ~ 4.6 kbp, so the blue indicated bands were cut out. ]] | wanted fragment size is ~ 4.6 kbp, so the blue indicated bands were cut out. ]] | ||
| Line 100: | Line 100: | ||
==== CPEC Assembly ==== | ==== CPEC Assembly ==== | ||
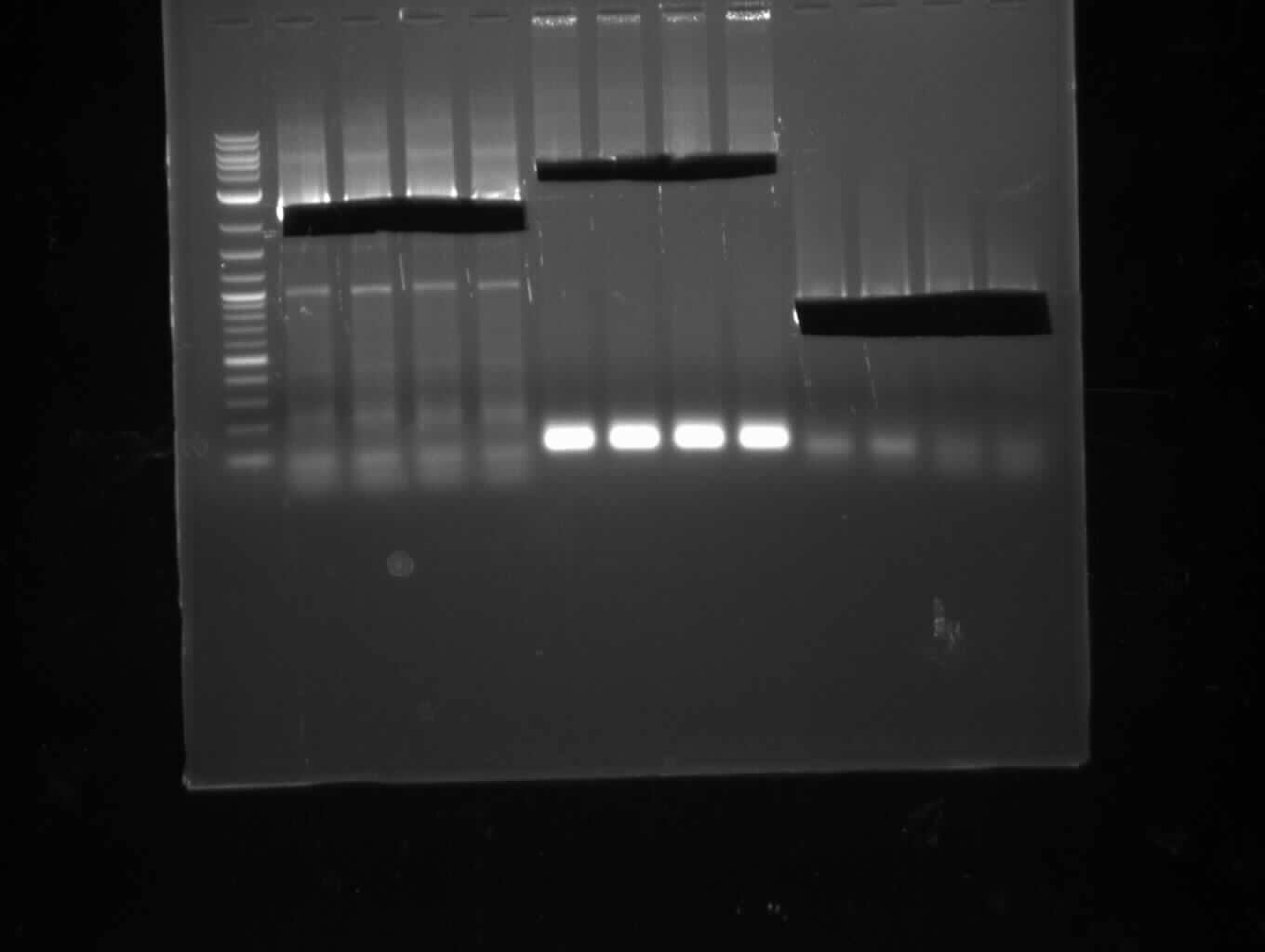
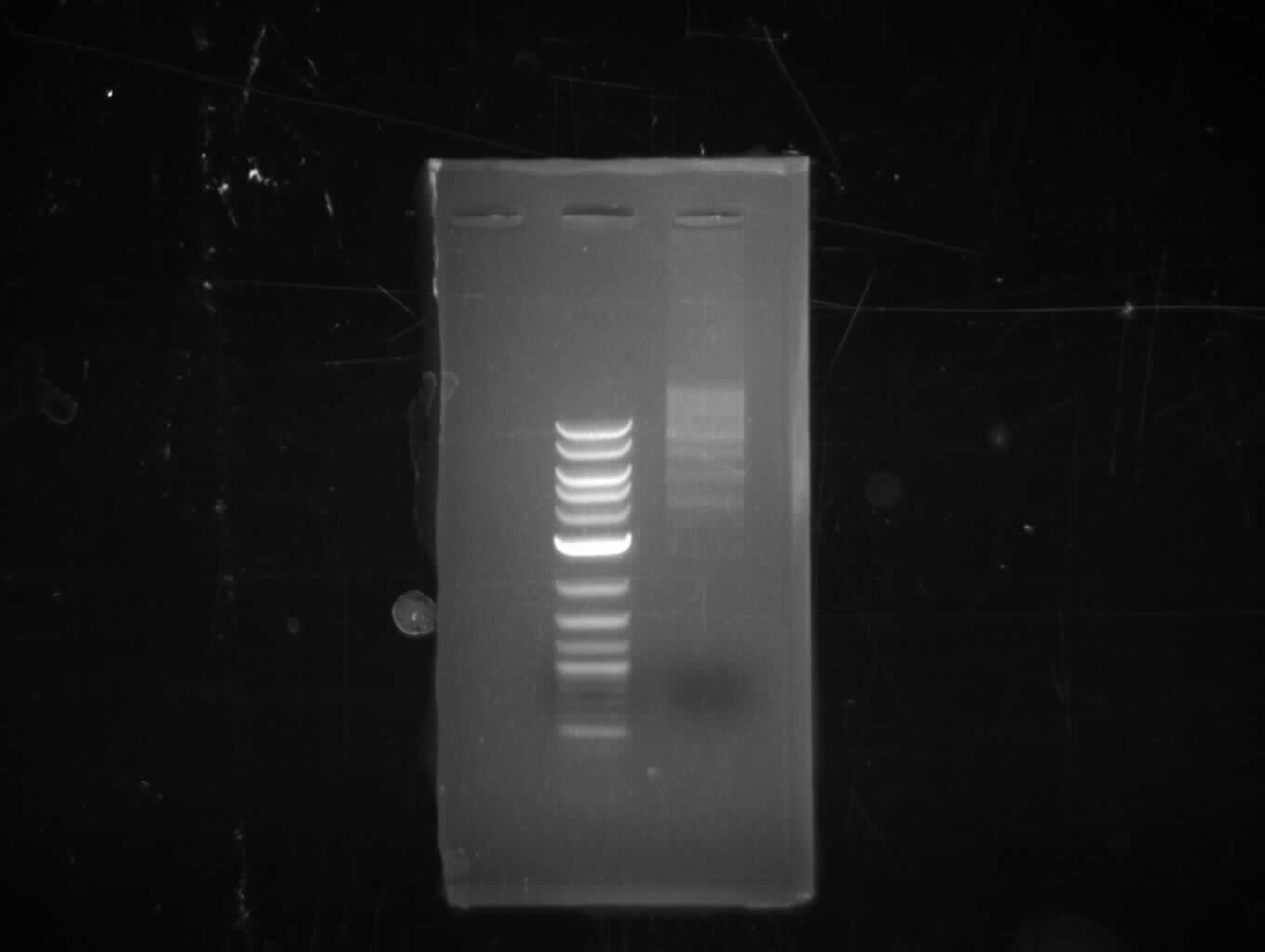
| - | [[File: | + | [[File:Heidelberg_20130713-CPEC.png|100px|thumb|right|CPEC for pKH1 assembly: only insert seems visible, no backbone and CPEC |
product; run at 135 V, 0.8 % gel (TAE); wanted amplicon size is: 2,4 kbp]] | product; run at 135 V, 0.8 % gel (TAE); wanted amplicon size is: 2,4 kbp]] | ||
| Line 140: | Line 140: | ||
==== PCR amplification f7 (pSB1C3) ==== | ==== PCR amplification f7 (pSB1C3) ==== | ||
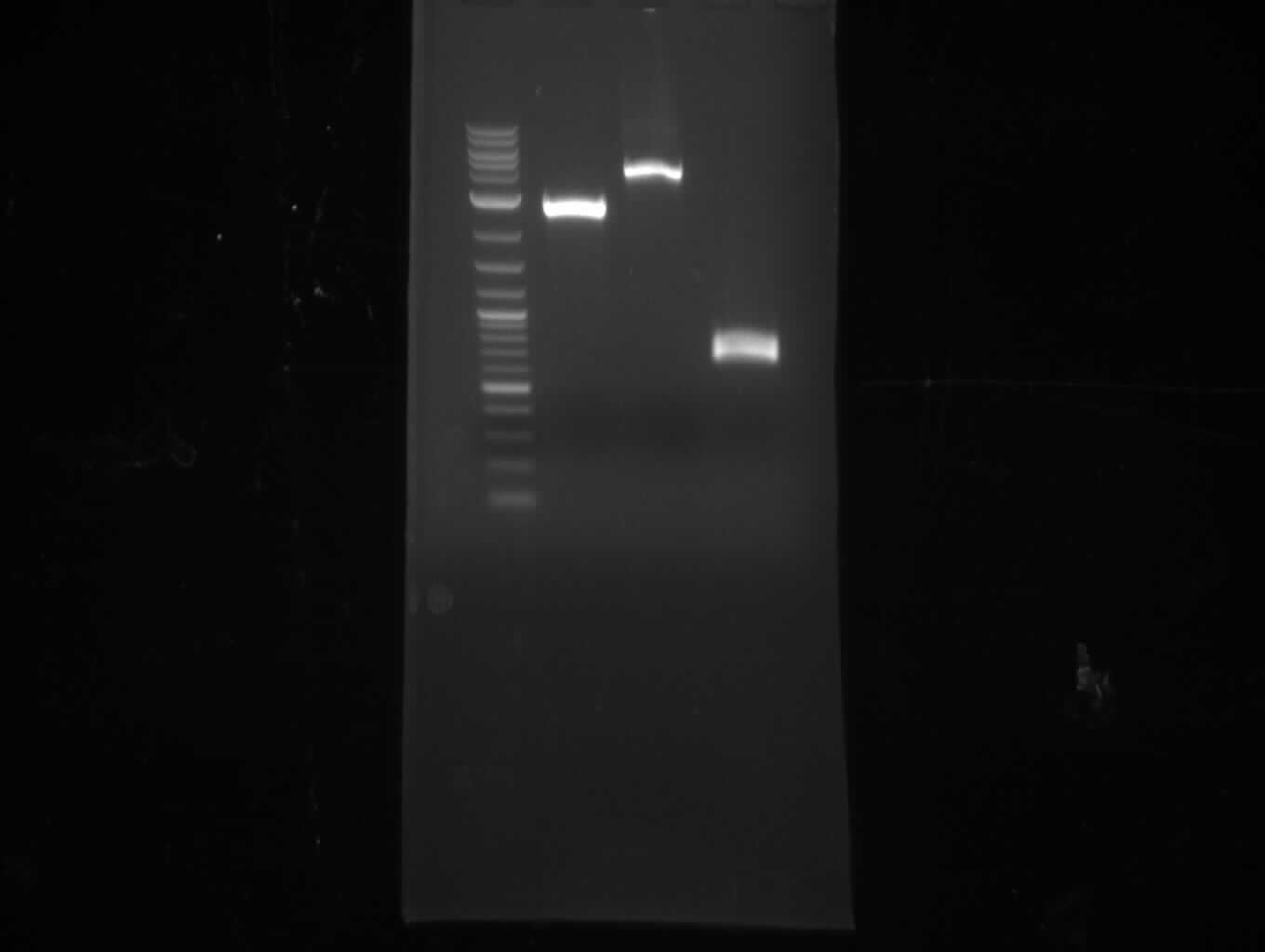
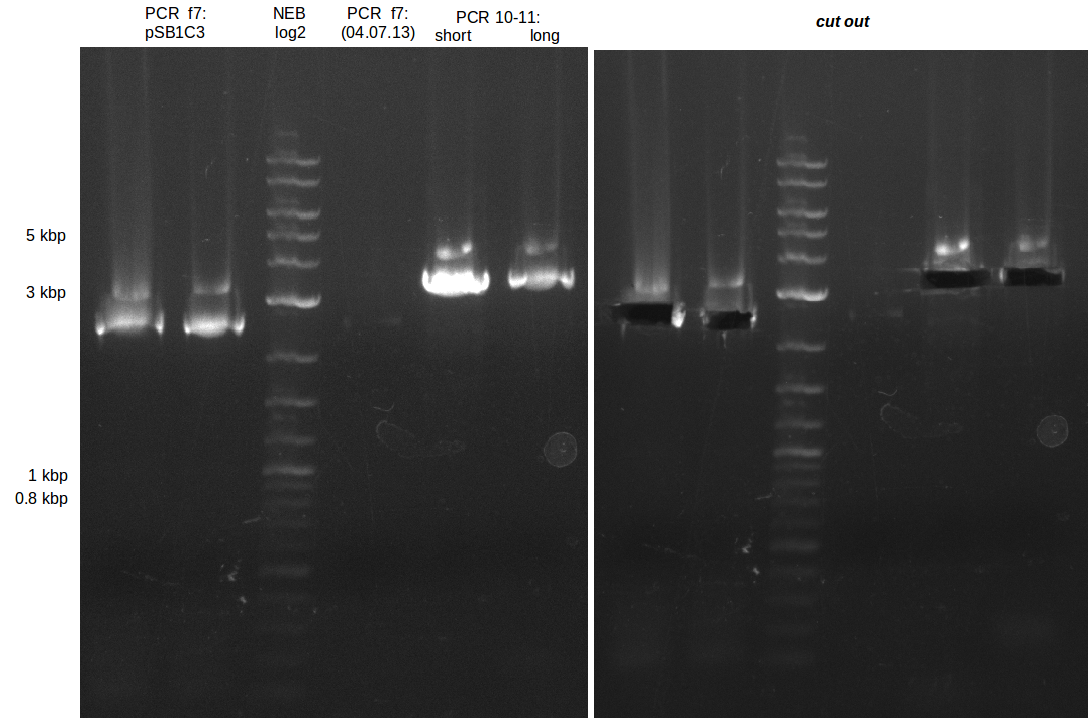
| - | [[File: | + | [[File:Heidelberg_20130713-f7-delRest.png|500px|thumb|right|PCR for amplification of pSB1C3: ('''left''') lane 1 & 2: todays |
amplification, lane 3: Marker, lane 4: 2 µl of gel/nucleotide purified amplification of 4th of July, lane 5 & 6: | amplification, lane 3: Marker, lane 4: 2 µl of gel/nucleotide purified amplification of 4th of July, lane 5 & 6: | ||
| Line 258: | Line 258: | ||
10 ul quickload 2-log; 2 ul PCR pSB1C3-indC-sfp; 29 ul 2x pSB1C3; 2x indC; 2x sfp | 10 ul quickload 2-log; 2 ul PCR pSB1C3-indC-sfp; 29 ul 2x pSB1C3; 2x indC; 2x sfp | ||
:<gallery widths="400px" heights="300px"> | :<gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130719_indigo1.jpg|400px|1st run with long primers | |
| - | + | File:Heidelberg_20130719_indigo1_cut.jpg|400px|cut backbone for gel extraction | |
</gallery> | </gallery> | ||
indC-Plum #2 | indC-Plum #2 | ||
| Line 314: | Line 314: | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130719_indigo2.JPG|2-log (1 ug); (1 ul) 1 - 2 - 3 - a - b; (all) 1 - 3 - 3 - a - b - b | |
| - | + | File:Heidelberg_20130719_indigo2_cut.JPG|2-log (1 ug); (1 ul) 1 - 2 - 3 - a - b; (all) 1 - 3 - 3 - a - b - b | |
</gallery> | </gallery> | ||
| Line 353: | Line 353: | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130719_sfpAF.JPG|2-log (1 ug); A-F | |
</gallery> | </gallery> | ||
| Line 445: | Line 445: | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130720_RB3-gibson.JPG|2-log (1 ug); pSB1C3; indC; sfp | |
| - | + | File:Heidelberg_20130720_RB3-gibson_cut.JPG|2-log (1 ug); pSB1C3; indC; sfp | |
</gallery> | </gallery> | ||
| Line 456: | Line 456: | ||
**pSB1C3 NheI-HF and KpnI-HF | **pSB1C3 NheI-HF and KpnI-HF | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130720_RB1-digest.jpg | |
| - | + | File:Heidelberg_20130720_RB1-digest_cut.JPG | |
</gallery> | </gallery> | ||
*Ligation 1 20 ul (2.0 T4 buffer 10x; 1.0 T4; 10.0 indC; 4.0 sfp; 3.0 pSB1C3) | *Ligation 1 20 ul (2.0 T4 buffer 10x; 1.0 T4; 10.0 indC; 4.0 sfp; 3.0 pSB1C3) | ||
| Line 464: | Line 464: | ||
** 2.0 T4 buffer 10x; 1.0 T4; 10.0 product; 7.0 pSB1C3 | ** 2.0 T4 buffer 10x; 1.0 T4; 10.0 product; 7.0 pSB1C3 | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_20130720_RB3-ligation.JPG | |
</gallery> | </gallery> | ||
| Line 495: | Line 495: | ||
<gallery widths="400px" heights="300px"> | <gallery widths="400px" heights="300px"> | ||
| - | + | File:Heidelberg_TOP10-pRB3-CPEG50-5ul.JPG|TOP10-pRB3-CPEG50-5ul | |
| - | + | File:Heidelberg_TOP10-pRB3-CPEG55-5ul.JPG|TOP10-pRB3-CPEG55-5ul | |
| - | + | File:Heidelberg_TOP10-pRB3-CPEG55lig-5ul.JPG|TOP10-pRB3-CPEG55lig-5ul | |
| - | + | File:Heidelberg_TOP10-pRB3-CPEG60-5ul.JPG|TOP10-pRB3-CPEG60-5ul | |
| - | + | File:Heidelberg_TOP10-pRB3-lig1-2ul.JPG|TOP10-pRB3-lig1-2ul | |
| - | + | File:Heidelberg_TOP10-pRB3-lig1-4ul.JPG|TOP10-pRB3-lig1-4ul | |
| - | + | File:Heidelberg_TOP10-pRB3-lig2-4ul.JPG|TOP10-pRB3-lig2-4ul | |
</gallery> | </gallery> | ||
Latest revision as of 00:46, 5 October 2013
Contents |
Indigoidine production pKH and pMM (Konrad)
IPTG Induction
Occulate 2 ml LB + appropiate AB with:
- Rosetta + pMM64 + pMM65
- BAP1
- BAP1 + pMM64
- BAP1 + pMM64 + pMM65
Rosetta was oculated from plate (06.07.), the BAP1 culture with 10 µl of ON from 06.07. These DC were incubated for
around 7 hours at 37 °C.
Old broth from 06.07 were induced with 1 mM IPTG at 24 °C for 2 hours. Rosetta strain becomes very blue, even
without IPTG induction.
fusion PCR (bpsA+svp)
based on PCA protocol: Used two main steps, first elongation of fragments (f1;f6) without any primers, than
enrichment of desired fusion product with fw primer of fragment 1 and rv primer of fragment 2.
Step 1:
- 13 µl H2O
- 25 µl Phusion Flash MM
- 2 µl template (f6:svp)
- 10 µl template (f1:bpsA from 4.07.)
(concentration of f1 from 4.07. unknown)
Step 2:
- 23.5 µl H2O
- 25 µl Phusion Flash MM
- 0.5 µl template (reaction from step 1)
- 2x 0.5 µl primer (NI:01; NI:08)
| Step | Cycles | temperature [°C] | Time [min] |
|---|---|---|---|
| 1 | 1 | 98 | 0:10 |
| 5 | 98 | 0:01 | |
| 52 | 0:30 | ||
| 72 | 0:20 | ||
| 1 | 72 | 5:00 | |
| 1 | 12 | inf | |
| use only 0.5 µl of mixture for 2nd step | |||
| 2 | 1 | 98 | 0:10 |
| 30 | 98 | 0:01 | |
| 72 | 1:10 | ||
| 1 | 72 | 5:00 | |
| 1 | 12 | inf | |
- Gel extraction of ~4.6 kbp bands cut out
Gibson assembly
- molecular ratio = backbone : inserts = 1 : 3
- partslength: bpsA=3.8 kbp; svp=0.8 kbp; pSB1C3=2.4 kbp
| fragment | volume [µl] | DNA amount [ng] |
|---|---|---|
| f8: bpsA + svp | 7.5 | 214 |
| f7: pSB1C3 (linear.) | 2.5 | 55 |
| total | 10 | |
| 0 µl | H2O | |
| 10 µl | Gibson MM |
- transformation of TOP10 with 2 µl directly of GA reaction mix and 1:4 dilution respectively
additional
- preparations of JM1 from 4.5 ml TOP10 ON culture (used a bit for transformation, gave rest to Johanna)
- transformation of BAP1 with 88 ng (5 µl) JM1 (probably way to much DNA)
- IPTG induction (1mM) experiment with Rosetta + pMM64 + pMM65, BAP1, BAP1 + pMM64, BAP1 + pMM64 + pMM65 (DC, was
in 4 °C fridge overnight)
- RESLUT: did not turn blue at all, may the IPTG be bad? (did not use the same as the day before)
CPEC Assembly
| fragment | volume [µl] | DNA amount [ng] |
|---|---|---|
| f8: bpsA + svp | 2.3 | 150 |
| f7: pSB1C3 (linear.) | 5.2 | 50 |
| total | 7.5 | |
| 2.5 µl | H2O | |
| 10 µl | PhusionFlash MM | |
| total | 20 |
| Cycles | temperature [°C] | Time [min] |
|---|---|---|
| 1 | 98 | 0:10 |
| 3 | 98 | 0:01 |
| 55 | 0:30 | |
| 72 | 2:00 | |
| 1 | 72 | 5:00 |
| 1 | 12 | inf |
- use 10 µl for gel picture, store 10 µl
PCR amplification f7 (pSB1C3)
(add items in this order)
- 14.6 µl H2O
- 25 µl Phusion MM
- 2x 0.5 µl Primer (NI09,NI10)
- 0.4 µl template (pSB1C3 with J04450 (pJM03))
| Cycles | temperature [°C] | Time [min] |
|---|---|---|
| 1 | 98 | 0:10 |
| 30 | 98 | 0:01 |
| 72 | 0:40 | |
| 1 | 72 | 5:00 |
| 1 | 12 | inf |
Ralf
new primers are there yippie
pSB1C3 RB21/22
- 2,5 ul template pSB1C3 1:5
- 25 ul Phusion Flash MM
- 5 ul Primer 10 mM
- 12,5 ul water
| 1 | 98 | 10 |
| 1 | 98 | 1 |
| 61 | 5 | |
| 72 | 60 | |
| 29 | 98 | 1 |
| 72 | 60 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
T1000 BIORAD
indC-Plum RB27/28
- 1 ul template cell pellet from DSMZ stem culture
- 25 ul Phusion Flash MM
- 5 ul Primer 10 mM
- 14 ul water
| 1 | 98 | 120 |
| 1 | 98 | 1 |
| 57 | 5 | |
| 72 | 90 | |
| 29 | 98 | 1 |
| 65 | 5 | |
| 72 | 90 | |
| 1 | 72 | 300 |
| 1 | 10 | - |
biometra
sfp-BAP1 RB35/36
- 1 ul template cell pellet from BAP1 liquid culture
- 25 ul Phusion Flash MM
- 5 ul Primer 10 mM
- 14 ul water
| 1 | 98 | 120 |
| 1 | 98 | 1 |
| 63 | 5 | |
| 72 | 30 | |
| 29 | 98 | 1 |
| 65 | 5 | |
| 72 | 30 | |
| 1 | 72 | 300 |
| 1 | 10 | - |
BioRAD 2-block
Electrophoresis 1 % Agarose 0,5 % TAE 100 V 60 min; PCR product 50 ul + 10 ul 6x loading dye 10 ul quickload 2-log; 2 ul PCR pSB1C3-indC-sfp; 29 ul 2x pSB1C3; 2x indC; 2x sfp
indC-Plum #2 Q5 50 ul 1, 2: RB37/38; 3: RB27/28
- 5 ul template liquid culture - pellet
- 25 ul Q5 MM
- 5 ul Primer 10 mM
- 10 ul water
cycles temperature °C time seconds 1 98 180 30 98 8 56 10 72 150 1 72 300 1 12 - sfp BAP1 #2 Q5 50 ul a: RB35/36 b: RB43/44
- template colony pick
- 25 ul Q5 MM
- 5 ul Primer 10 mM
- 15 ul water
cycles temperature °C time seconds 1 98 180 30 98 8 62 10 72 40 1 72 300 1 12 - analysis
- indC fragmet with short primers worked well -> gel extraction and re-PCR with long primers
- sfp and indC long almost nothing -> improve conditions
sfp BAP1 #3 Q5 50 ul A: RB17/18; naka; 60 °C B: RB17/18; naka; 56 °C C: RB43/44; short; 60 °C D: RB43/44; short; 56 °C E: RB35/36; long; 60 °C F: RB35/36; long; 65 °C
- template colony pick
- 25 ul Q5 MM
- 5 ul Primer 10 mM
- 15 ul water
cycles temperature °C time seconds 1 98 180 30 98 8 56-60-65 10 72 30 1 72 300 1 12 - analysis
- B, C and E worked best -> gel extraction
- re-pcr E
fragments for pRB3
- rePCR from indC-Plum #2 with long primers
- 2x 50 ul Phusion Flash HF
- RB27/28 long primer for pRB3 5 ul 10 uM
- water 14.8 ul
- template 0.2 ul pcr gel extracted
biorad T100
cycles temperature °C time seconds 1 98 10 12 98 1 td 57-53 -0.5 5 72 70 18 98 1 65 5 72 70 1 72 300 1 12 - - rePCR from sfp-BAP1 #3 E with long primers
- 2x 50 ul Phusion Flash HF
- RB35/36 long primer for pRB3 5 ul 10 uM
- water 14.8 ul
- template 0.2 ul pcr gel extracted
biorad T100
cycles temperature °C time seconds 1 98 10 30 98 1 65 5 72 20 1 72 300 1 12 - - rePCR from pSB1C3 Hanna 1:5 with long primers
- 2x 50 ul Phusion Flash HF
- RB21/22 long primer for pRB3 5 ul 10 uM
- water 14.8 ul
- template 0.2 ul pcr gel extracted
cycles temperature °C time seconds 1 98 10 12 98 1 td 64 -0.5 5 72 70 23 98 1 65 5 72 70 1 72 300 1 12 -
assembly pRB3
- digest 25 ul (2.5 CutSmart 10x; 0.5 enzyme each; 21.5 DNA; 1 h at 37 °C)
- indC KpnI-HF and BamHI-HF
- sfp BamHI-HF and NheI-HF
- pSB1C3 NheI-HF and KpnI-HF
- Ligation 1 20 ul (2.0 T4 buffer 10x; 1.0 T4; 10.0 indC; 4.0 sfp; 3.0 pSB1C3)
- Ligation 2
- 2.0 T4 buffer 10x; 1.0 T4; 7.0 indC; 10.0
- 2.0 T4 buffer 10x; 1.0 T4; 10.0 product; 7.0 pSB1C3
- CPEG 20 ul Phusion Flash HF (10 MM; 7 indC; 2 pSB1C3; 1 sfp)
biometra
cycles temperature °C time seconds 1 98 10 10 98 1 3grad. 50 +5 5 72 70 1 72 180 1 12 - - Transformation TOP10
- CPEG 50 5 ul
- CPEG 55 5 ul
- CPEG 55 plus 20 min T4 ligase 10 ul (5 ul + 5 ul ligation master mix)
- CPEG 60 5 ul
- Lig1 2 ul
- Lig1 4 ul
- Lig2 4 ul
-> order primers for the other PPTases and indigoidine synthetases (set RB21-44 complete)
Results and Discussion
 "
"