Team:TU-Eindhoven/Preparation
From 2013.igem.org
JacquesErnes (Talk | contribs) (→Protein Design) |
JacquesErnes (Talk | contribs) (→Protein Design) |
||
| Line 29: | Line 29: | ||
The proteins we will use are mostly consisting of repeating sequences with large numbers of Arginine and Lysine amino acids. These repeating sequences were chosen because of their good CEST qualities {{:Team:TU-Eindhoven/Template:Ref| id=McMahonDIACEST | author=M.T. McMahon | title=New "Multicolor" Polypeptide Diamagnetic Chemical Exchange Saturation Transfer (DIACEST) Contrast Agents for MRI | journal=Magnetic Resonance in Medicine | edition=60 | pages=803-812 | year=2008 }}. | The proteins we will use are mostly consisting of repeating sequences with large numbers of Arginine and Lysine amino acids. These repeating sequences were chosen because of their good CEST qualities {{:Team:TU-Eindhoven/Template:Ref| id=McMahonDIACEST | author=M.T. McMahon | title=New "Multicolor" Polypeptide Diamagnetic Chemical Exchange Saturation Transfer (DIACEST) Contrast Agents for MRI | journal=Magnetic Resonance in Medicine | edition=60 | pages=803-812 | year=2008 }}. | ||
| - | Of the 10 proteins 4 were pre-existing proteins and did not need any adaptations (although being rather short we linked multiple repeats of these proteins together.) However, selecting these naturally occurring proteins did cost some effort. A program was written to asses the properties of many hundreds of proteins and those with the best attributes were chosen for use in the lab. The exact method of selection is explained [[Team:TU-Eindhoven/ProteinSelection | here]]. The remaining 6 proteins were not naturally occurring proteins, and were based on a repeating sequence of either 2, or 4 amino acids. As these repeating sequences all used the same amino acids it was of utmost importance to vary the codon sequence as best we could whilst also adhering to the E.coli occurrence frequencies of each codon. To help achieve this goal, a small program was written in Python. Here the E.coli occurrence frequency of each codon was used to randomize the protein sequence whilst maintaining a low Tm for the entire protein and reducing the risk of hairpin forming and dimer forming within the protein. This way we managed to find the in our eyes optimal DNA sequence for both the DNA synthesis and the protein expression later on. | + | Of the 10 proteins 4 were pre-existing proteins and did not need any adaptations (although being rather short we linked multiple repeats of these proteins together.) However, selecting these naturally occurring proteins did cost some effort. A program was written to asses the properties of many hundreds of proteins and those with the best attributes were chosen for use in the lab. The exact method of selection is explained [[Team:TU-Eindhoven/ProteinSelection | here]]. The remaining 6 proteins were not naturally occurring proteins, and were based on a repeating sequence of either 2, or 4 amino acids. As these repeating sequences all used the same amino acids it was of utmost importance to vary the codon sequence as best we could whilst also adhering to the E.coli occurrence frequencies of each codon. To help achieve this goal, a small program was written in Python. Here the E.coli occurrence frequency of each codon was used to randomize the protein sequence whilst maintaining a low Tm for the entire protein and reducing the risk of hairpin forming and dimer forming within the protein. This way we managed to find the, in our eyes, optimal DNA sequence for both the DNA synthesis and the protein expression later on. |
====Final Design==== | ====Final Design==== | ||
Latest revision as of 12:15, 26 October 2013



Preparation
Before a start could be made on the lab work it was of course of importance that detailed protocols and plannings were made alongside other preperations. The aim of this page is to provide an overview of these initial steps leading up to the lab work.
Contents |
Construct Design
Before commencing with the lab-work, DNA constructs, which will form the basis of all our lab work, were designed using GenomeCompiler and ordered at GeneScript. As these constructs will form the basis for such a large part of our lab-work it seemed logical for us to go into the details of their design.
General Design
In the lab we wished to compare a number of different proteins which led to the design of a general construct in which only the specific protein sequence would need to be adapted for each of the proteins we wished to test. The general construct can be regarded as follows:
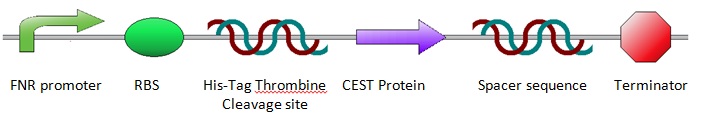
The general construct was designed in this way to enable us to use this construct for almost all lab experiments meaning no other constructs would have to be designed and ordered separately. We will use the EcoRI restriction site in the official iGEM restriction site (which is placed in front of the FNR promoter) in combination with the HindIII restriction site (behind the terminator and behind the official iGEM restriction sites) to transfer the entire construct from the pUC57-Simple vector in which the construct has been ordered to the pBR322 vector in which we can perform anaerobic experiments, testing the FNR promoter. We can use the NheI (after the Thromine cleavage site) and XhoI (behind the variable protein sequence) restriction sites to transfer the protein sequence from the pUC57-Simple vector to the standard pET28a expression vector. Within the pET28a vector the proteins will be brought to expression aerobically allowing for protein purification, quantification and finally CEST imaging. Other parts of the general construct such as the His Tag, Thrombine Cleavage Site and the Terminator sequence have been included to optimize transcription and ultimately expression.
Protein Design
We will be testing the CEST contrast of 10 different proteins during the course of this project. Alongside this the fluorescent protein EGFP will be used as a control protein for all the experiments. The proteins we will use are mostly consisting of repeating sequences with large numbers of Arginine and Lysine amino acids. These repeating sequences were chosen because of their good CEST qualities McMahonDIACESTM.T. McMahon, New "Multicolor" Polypeptide Diamagnetic Chemical Exchange Saturation Transfer (DIACEST) Contrast Agents for MRI. Magnetic Resonance in Medicine 60, 803-812 (2008).
Of the 10 proteins 4 were pre-existing proteins and did not need any adaptations (although being rather short we linked multiple repeats of these proteins together.) However, selecting these naturally occurring proteins did cost some effort. A program was written to asses the properties of many hundreds of proteins and those with the best attributes were chosen for use in the lab. The exact method of selection is explained here. The remaining 6 proteins were not naturally occurring proteins, and were based on a repeating sequence of either 2, or 4 amino acids. As these repeating sequences all used the same amino acids it was of utmost importance to vary the codon sequence as best we could whilst also adhering to the E.coli occurrence frequencies of each codon. To help achieve this goal, a small program was written in Python. Here the E.coli occurrence frequency of each codon was used to randomize the protein sequence whilst maintaining a low Tm for the entire protein and reducing the risk of hairpin forming and dimer forming within the protein. This way we managed to find the, in our eyes, optimal DNA sequence for both the DNA synthesis and the protein expression later on.
Final Design
The combination of all the above mentioned parts into a single DNA sequence turned out to be a timely and often complex task. Whilst this task was made somewhat easier by the use of the GenomeCompiler a number of weeks passed before we had managed to fully finalize the DNA sequences. These DNA sequences can be opened in the following link, with a color coding to help you analyze each of the separate parts:
References
 "
"



