Team:HokkaidoU Japan/Shuffling Kit/Examples
From 2013.igem.org
| Line 29: | Line 29: | ||
<div class="fig fig800"> | <div class="fig fig800"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | <img src="https://static.igem.org/mediawiki/2013/2/2d/Fig2_in_example_HokkaidoU_2013.png"> |
<div style="padding-bottom: 0;"><span class="bold">fig.2 Difference of Kanamycin concentration.</span></div> | <div style="padding-bottom: 0;"><span class="bold">fig.2 Difference of Kanamycin concentration.</span></div> | ||
</div> | </div> | ||
Revision as of 12:47, 27 October 2013
Maestro E.coli
Optimization Kit

Demonstrations for Usecase Example
We will show some interesting demonstrations of our kits, Promoter Selector and RBS Selector!
Promoter Selector
Let's select the best promoter for Kanamycin resistance by Promoter Selector.
For a demonstration we decided to optimize the expression of Kanamycin resistance. Changing the concentration of Kanamycin in agar plate, it is estimated that different promoter will be chosen by our Promoter Selector (fig.1).
If the concentration of Kanamycin was high, the colony with strong promoter will survive. Therefore, only one or two colors of colonies would appear. If the concentration of Kanamycin was low, colonies with weak promoters will be able to survive. This way many colors of colonies would appear (fig.2).


Method
Optimum concentration of Kanamycin: in LB is 50 mg/ml We prepared 3 different concentration plates.
- Plate A: Kanamycin 125 mg per plate
- Plate B: Kanamycin 250 mg per plate
- Plate C: Kanamycin 500 mg per plate (optimum concentration)
- Plate D: Kanamycin 1000 mg per plate
Gene Vector: pSB1C3
We cloned Kanamycin resistant gene from pSB1K3, by using BsaI adding primer. Used the Promoter Selector (K1084501, K1084502, K1084503, K1084504, K1084505 ).
Culture: 37 °C, for 24h

Results
We didn't have time to culture anymore. So we coudn't see the diffference between plates A to D. However,we were able to see different colors on the plate. The colonies were too small to identify the color. The picture below is plate B.
Conclusion
Our Promoter Selector seemed to work. We got several colors colonies on Kanamycin plates. This means that Kanamycin resistance gene was successfully ligated. We need more than 24 hrs to identify the colors.
RBS Selector
4 colors
Let’s create all combinations by two reporter genes and make various colors on one plate!
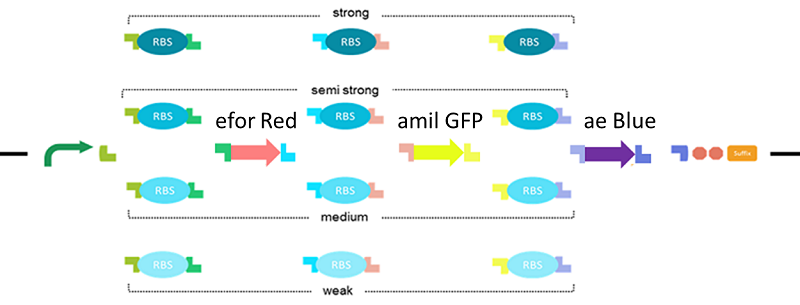
The RBS Selector we made, can randomize the strength of RBSs in the operon. For a demonstration, we decided to create all combinations by two genes; mRFP1 (BBa_E1010) and LacZα (BBa_I732006) (fig.4). LacZα makes the colony blue. mRFP1 makes the colony red.

When the RBS upstream of mRFP1 was strong and the RBS upstream was weak, the colony should be red. When the RBS upstream of mRFP1 was weak, and the RBS upstream was strong, the colony should be blue. So when if the strength of RBS upstream both genes were the same, colony will be white, purple (fig.5).

Method
- Used promoter1 (BBa_K1084001), SD2 (BBa_K1084101), SD4 (BBa_K1084102) and assembled with.
- Spread X-GAL(250 mg)on LBC plate.
- Cultured for 37 °C, 26h.

Results
We got many colored colonies,red, blue, white, and purple.
Conclusion
We can say that our RBS Selector worked!! The RBSs uptsream 2 genes were randomized and they had many levels of expressions.
64 colors
 "
"


