Team:DTU-Denmark/Notebook/24 July 2013
From 2013.igem.org
(→208) |
|||
| Line 4: | Line 4: | ||
==Main purpose== | ==Main purpose== | ||
<hr/> | <hr/> | ||
| + | Make USER-reaction to get pZA21 with arabinose inducible promoter. Make PCR to get more HAO and AMO with USER-primers. Finish the restriction analysis. | ||
==Who was in the lab== | ==Who was in the lab== | ||
| Line 62: | Line 63: | ||
==Results== | ==Results== | ||
| - | ==first gel== | + | ===first gel=== |
[[File:2013-07-24 nopro colony extraction.jpg|600px]] | [[File:2013-07-24 nopro colony extraction.jpg|600px]] | ||
| Line 68: | Line 69: | ||
decided to purify RFP in pZA21 without promoter as well as HAO, AMO and cycAX | decided to purify RFP in pZA21 without promoter as well as HAO, AMO and cycAX | ||
| - | ==purification gel== | + | ===purification gel=== |
loaded the complete PCR reaction (~45 uL), cut out the bands of our products and used QIAgen gel extraction kit | loaded the complete PCR reaction (~45 uL), cut out the bands of our products and used QIAgen gel extraction kit | ||
| Line 82: | Line 83: | ||
[[File:2013-07-24 purification.jpg|600px]] | [[File:2013-07-24 purification.jpg|600px]] | ||
| - | ==gel for restriction analysis== | + | ===gel for restriction analysis=== |
[[File:2013-07-24 restriction amo hao cyc.jpg|600px]] | [[File:2013-07-24 restriction amo hao cyc.jpg|600px]] | ||
Revision as of 20:00, 24 July 2013
22 July 2013
Contents |
208
Main purpose
Make USER-reaction to get pZA21 with arabinose inducible promoter. Make PCR to get more HAO and AMO with USER-primers. Finish the restriction analysis.
Who was in the lab
Henrike, Julia, Gosia, Kristian
Procedure
USER reaction and transformation
We perform USER reaction, samples are as follows:
- plasmid pZA21 and AMO
- plasmid pZA21 and HAO
- negative control with water instead of insert
Reaction is performed in the same way as on 18-07-2013 with increased amount of insert (up to 14 uL due to low DNA concentration).
With the same procedure we also preformed a USER reaction with pZA21::RFP PCR-amplified with no promoter and araBAD from BBa_K808000.
Restriction analysis
From last week transformants with AMO and HAO in pZA21 we performed plasmid isolation.
We perform restriction analysis with EcoRI. Expected fragments are as follows:
- For pZA21 with AMO:
3309 pz, 2283 pz
- For pZA21 with HAO:
2233 pz, 2124 pz, 930 pz
Primer diluting
PCR to extract Nir from Pseudosomonas with new USER primers
Tubes have 4 labels (pZA21 only has 3), in vertical order:
- 'Nir' or 'pZ', tells which product is being made
- '1' - part 1 of Nir OR '2' - part 2 of Nir OR 'w' - whole Nir (on the pZ tubes 1 and 2 are just duplicates)
- '5' - used 5uL of N.europeae culture as template OR '10' - used 10uL of N.europeae culture as template
- 'ex' - extraction PCR, using the new non-USER primers that Jakob made OR 'U' - using the new USER primers that Jakob made
two programs:
all extraction PCRs and part 2 of Nir with USER primers - 54C, 5:00
part 1 of Nir with USER primers and pZA21 for ligation with Nir - 50C, 5:00
PCR with USER-primers on HAO and AMO
Standard USER-PCR was made with the HAO and AMO templates purified earlier today. The reactions where done in duplicates and with 2 different concentrations:
- HAO 5 is with 5uL template
- HAO 10 is with 10uL template
- AMO 5 is with 5uL template
- AMO 10 is with 10uL template
All tubes where run with 52C annealing temperature and 3 min extension time.
Results
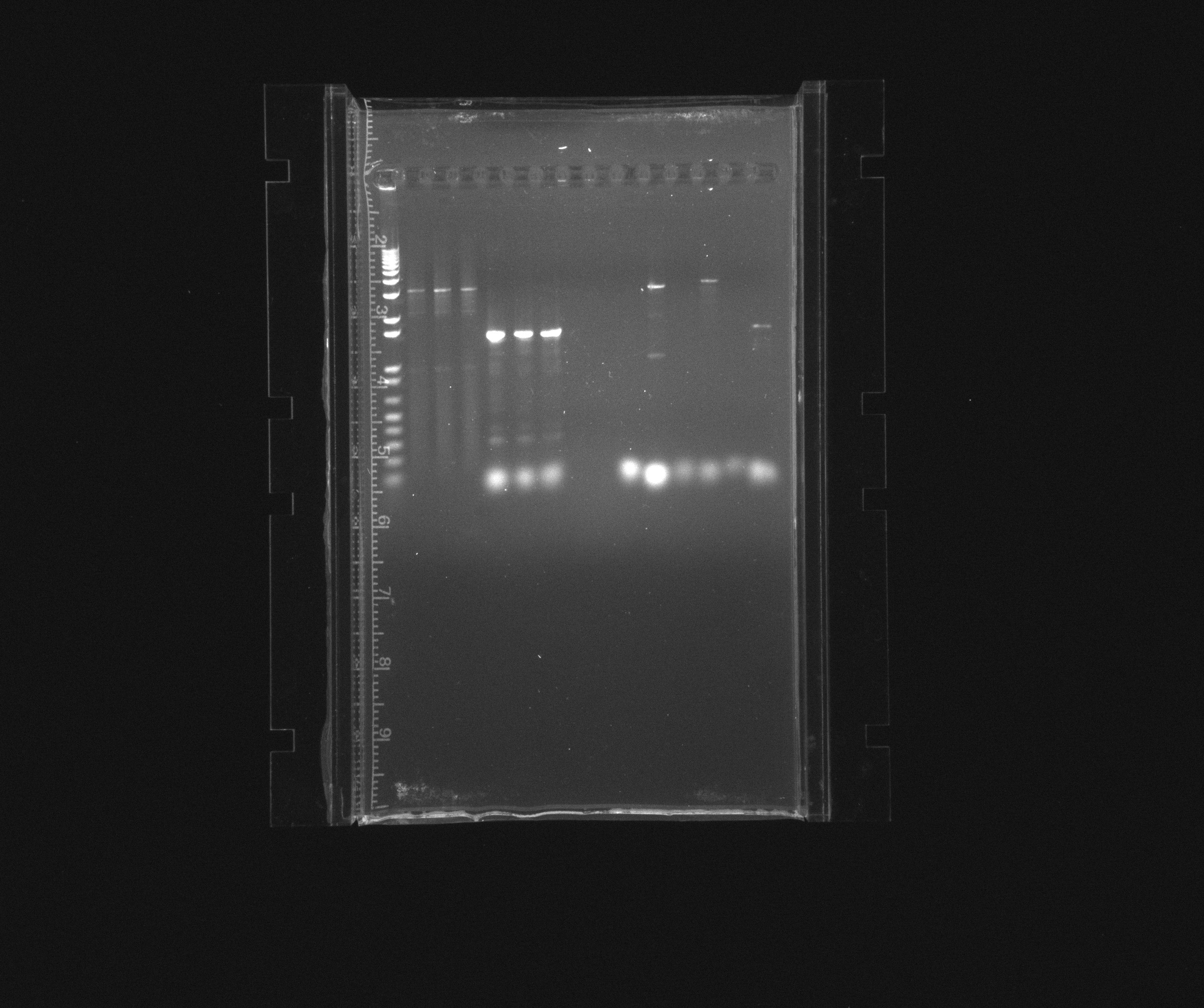
first gel
decided to purify RFP in pZA21 without promoter as well as HAO, AMO and cycAX
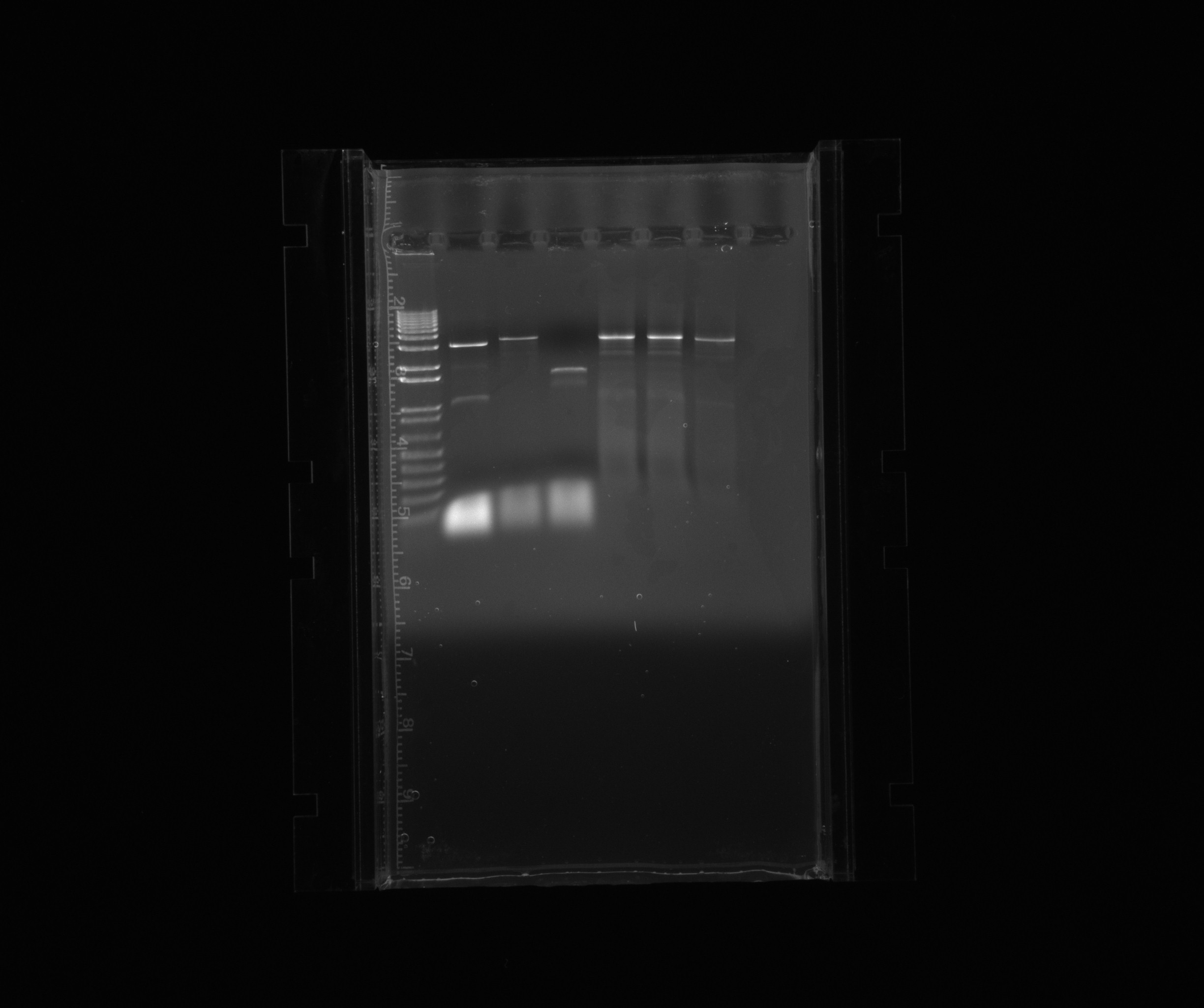
purification gel
loaded the complete PCR reaction (~45 uL), cut out the bands of our products and used QIAgen gel extraction kit
- 1: 1 kb ladder
- 2: HAO
- 3: AMO
- 4: cycAX
- 5: RPF in pZA21 without promoter
- 6: RPF in pZA21 without promoter
- 7: RPF in pZA21 without promoter
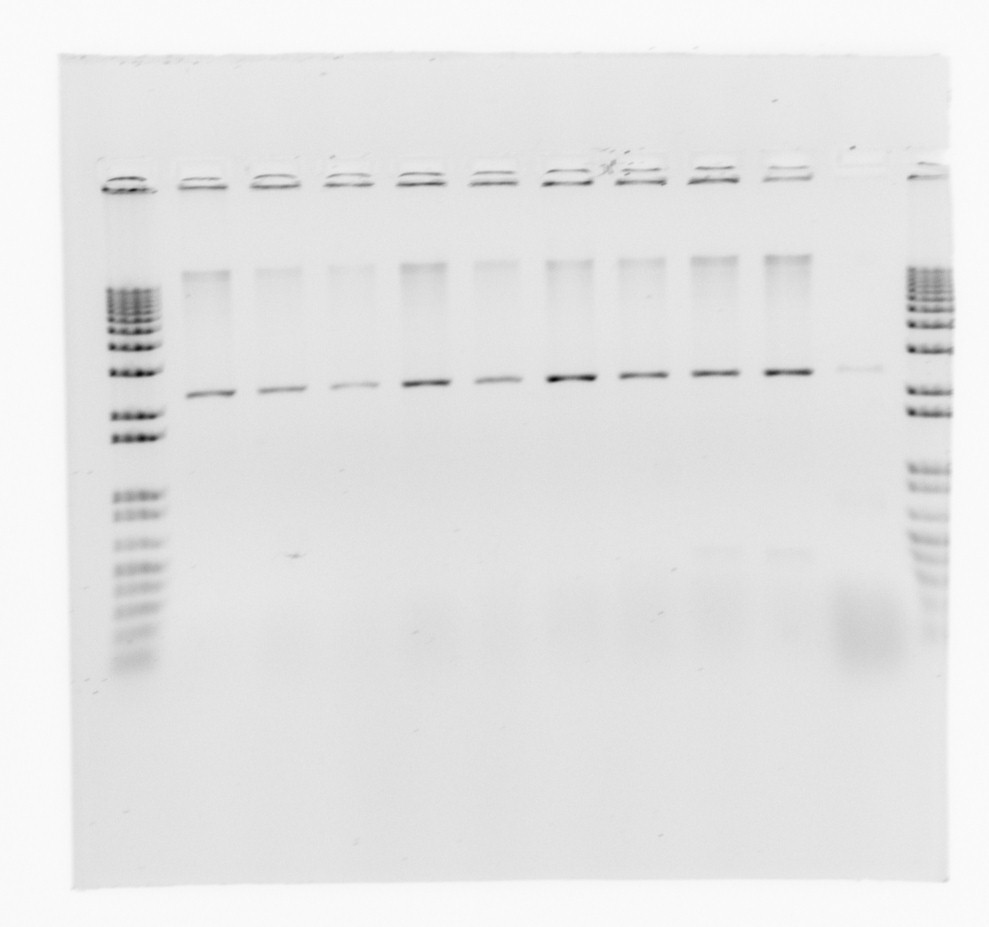
gel for restriction analysis
Conclusion: We only get one band for each plasmid, so the inserts are not present.
Conclusion
Navigate to the Previous or the Next Entry
 "
"