Team:DTU-Denmark/Notebook/22 July 2013
From 2013.igem.org
(→Results) |
|||
| Line 111: | Line 111: | ||
[[File:2013-07-22 cycax pza no prom.jpg|600px]] | [[File:2013-07-22 cycax pza no prom.jpg|600px]] | ||
| - | Navigate to the [[Team:DTU-Denmark/Notebook/ | + | Navigate to the [[Team:DTU-Denmark/Notebook/19_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/23_July_2013|Next]] Entry |
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Revision as of 10:42, 25 July 2013
22 July 2013
208
Main purpose
- ON culture of Colony PCR products of HAO and AMO from 19-07-2013
- USER reaction of AMO and HAO with pZA21 (with native promoter).
- RFP amplification in pZA21 using primers without promoter
- PCR to amplify USER-compatible AMO and HAO fragments
Who was in the lab
Gosia, Henrike, Kristian, Julia
Procedure
ON cultures of transformants
Innoculated ON cultures of the colonies from USER cloning of AMO and HAO on the 18.07. into 5mL LB medium. Will purify plasmids tomorrow.
USER reaction of AMO and HAO with pZA21 (with native promoter)
USER reaction was performed according to standard protocol but using more fragment because of the low DNA concentration in the PCR purification of the fragments (***).
USER mix:
- USER enzyme 1 uL
- NEB 4 buffer 0,5 uL
- 10x BSA 0,5 uL
- backbone pZA21 1uL
- Sample 1 -> 3 uL of USER mix + 12 uL of AMO
- Sample 2 -> 3 uL of USER mix + 12 uL of HAO
- Sample 3 -> Negative control, USER mix + 12 uL water
USER reaction was performed in PCR machine with programm: 37C for 40 min and then 25C for 30 min.
To 100uL of chemically competent E. coli cells 10 uL of USER mix after USER reaction was added.
Incubation of cells on ice for 30 minutes, heat shock at 42C for 90 sec. and 5 min on ice. Addition of SOC medium and 2 hours of incubation at 37C. Plating on plates with LB medium with Kanamycin, overnight incubation at 37C.
PCR to amplify pZA21 containing RFP without its native promoter
Since insertion of RFP into the vector pZA21 was confirmed by restriction analysis we will now amplify the plasmid without its native promoter and ligate the product with the USER fragment of the inducible araBAD promoter from BioBrick ??.
PCR settings standard with 57°C annealing temperature and 4 mins elongation time
PCR of AMO and HAO with USER endings
Performed PCR to extract AMO and HAO from nitrosomonas europeae as the amount of purified fragment is getting lower. Settings according to previously successful PCRs were:
AMO: 54°C, 3 mins
HAO: 56°C, 3 mins
Preparation of 1kb plus ladder from Invitrogen for use
200 uL of ladder were diluted in 800 uL of destilled water and 200 uL of loading buffer.
Perparation of stock solution of trace elements for DM medium
Made and autoclaved 200 mL 10x concentrated stock solution. Use only 1 mL in 1L medium instead of 10 mL.
Components:
2 mg MnCl2 tetrahydrate
27 mg FeCl3 hexahydrate
1.2 mg Na2MoO4 dihydrate
6.8 mg ZnSO4 heptahydrate
1.2 mg CoCl2 hexahydrate
Results
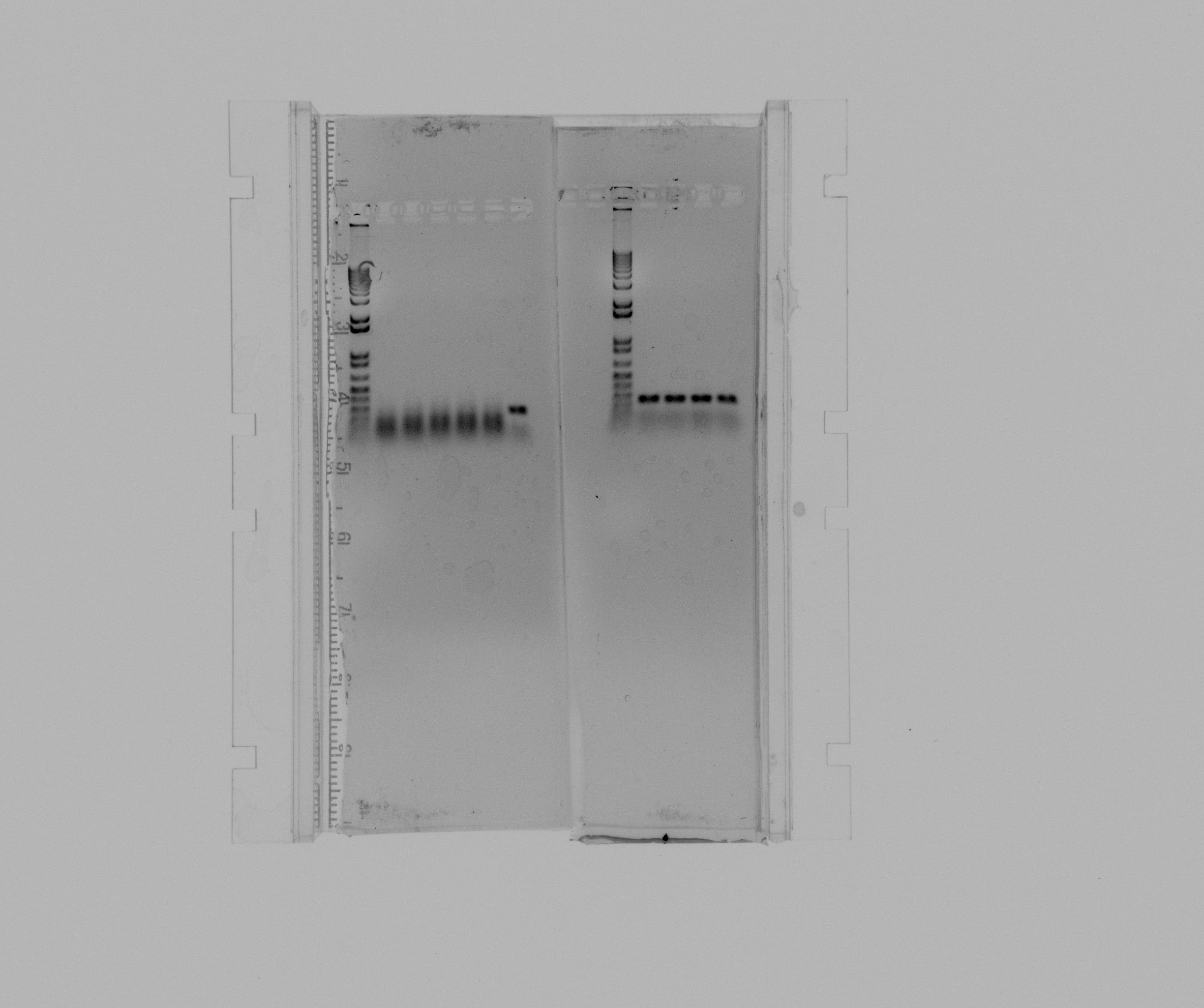
Run 1% agarose gel to analyze products on the PCR for AMO and HAO performed today. AMO is not observed and instead of HAO we receive an unknown fragment of length ~300 bp.
left:
- 1: 1kb ladder
- 2: AMO
- 3: AMO
- 4: AMO
- 5: AMO
- 6: AMO
- 7: HAO
right:
- 1: 1kb ladder
- 2: HAO
- 3: HAO
- 4: HAO
- 5: HAO
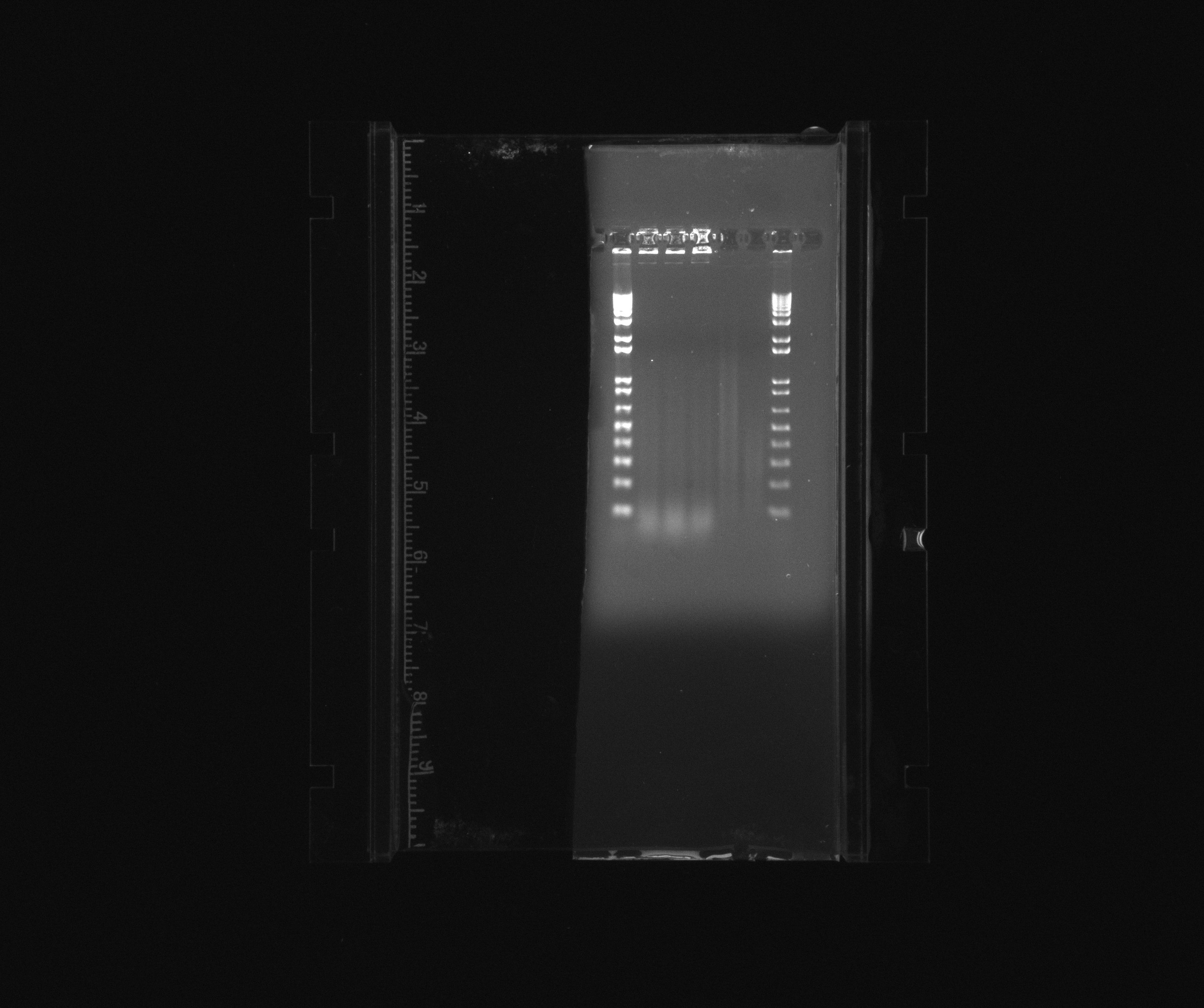
Run 1% gel on PCR for cycAX and the vector without promoter. Empty.
- 1: 1kb plus ladder 10uL
- 2: cycAX
- 3: cycAX
- 4: cycAX
- 5: pZA21 with RFP no promoter
- 6: pZA21 with RFP no promoter
- 7: 1kb plus ladder 5 uL
Navigate to the Previous or the Next Entry
 "
"