Notebook
Transformed BioBricks
Tuesday 18.06.13
Started transforming BioBricks. Transformed some composite BioBricks consisting of constitutive promotor, RBS, a fluorescent protein and a terminator. Transformed the parts Promoter+RBS+RFP+term (<partinfo>BBa_I13521</partinfo>), Promoter+RBS+GFP+term (<partinfo>BBa_I13522</partinfo>), Promoter+RBS+CFP+term (<partinfo>BBa_I13600</partinfo>) and Pm+RBS+mCherry+term to competent E.coli DH5α cells, to have some colorful cells to show the camera crew from NRK (Norwegian broadcasting), who will visit our lab tomorrow :-)
For our transformation protocol, see the protocol page.
Wednesday 19.06.13
We transformed BioBricks consisting of fluorescent proteins. Transformed the parts YFP (<partinfo>BBa_E0030</partinfo>), CFP (<partinfo>BBa_E0020</partinfo>), RFP (<partinfo>BBa_E1010</partinfo>), GFP (<partinfo>BBa_E0040</partinfo>), BFP (<partinfo>BBa_K592100</partinfo>) and SYFP (<partinfo>BBa_K864100</partinfo>).
We transferred mCherry from agar plate to liquid medium and incubated at 37° C.
Thursday 20.06.13
We transferred the transformed cells from yesterday to liquid medium and incubated at 37°C protocol.
We isolated the DNA from the transformed mCherry cells by using the miniprep protocol and measured the concentration by [http://www.nanodrop.com/library/nd-1000-v3.7-users-manual-8.5x11.pdf NanoDrop ND-1000 Spectrophotometer].
| Sample
| Concentration (ng/µl)
|
| mCherry1
| 5,6
|
| mCherry2
| 7,3
|
Friday 21.06.13
We isolated the DNA from the transformed cells by using the miniprep protocol and measured the concentration by [http://www.nanodrop.com/library/nd-1000-v3.7-users-manual-8.5x11.pdf NanoDrop ND-1000 Spectrophotometer].
| Sample
| Concentration (ng/µl)
|
| CFP1
| 8,59
|
| CFP2
| 7,04
|
| BFP
| 7,06
|
| SYFP
| 5,42
|
| RFP1
| 5,04
|
| RFP2
| 7,52
|
| YFP
| 6,79
|
| GFP
| 5,27
|
Monday 24.06.13 - Friday 28.06.13
This week we have established protocols for further work and designed primers for PCR.
Monday 01.07.13
We transformed some BioBricks: pBAD promoter (<partinfo>BBa_K206000</partinfo>), RBS (<partinfo>BBa_J61101</partinfo>) and a plasmid backbone (<partinfo>BBa_J01101</partinfo>).
Tuesday 02.07.2013
This day we started making the small-scale vesicle preparation according to our protocol.
Overnight cell cultures of E.coli ER2566 and E.coli BW27784 transformed with the pUM9 plasmid were preprared. The pUM9 plasmid has ampR induces a stress respons in E.coli when arabinose is present. According to the litterature (link?) stress in E.coli increases vesicle production.
The ER2566 cell were cultured in plain LB, while the BW27784 cells were cultured in LB with ampenicillin (100 µg/mL).
We transferred the samples of RBS, pBAD and the plasmid backbone (pTet) from agar plate to liquid medium and incubated at 37° C.
Wednesday 03.07.2013
The small-scale vesicle preparation continued with completion of step 2-7 in the vesicle isolation protocol. There where made three cell cultures in step 2; E.coli ER2566 in LB, E.coli BW27784 in LB with ampenicillin (100 µg/mL) (ara-) and E.coli BW27784 in LB with ampenicillin (100 µg/mL) with the addition of arabinose (0.5 %) after 1 hour of incubtion (ara+). The cell cultures in step 2 was incubated for 3 hours.
Optical denisty (OD) at 600 nm was mesured as indicated in the table below. No dilution of the samples was necessary.
| Sample
| OD600
|
| ER2566
| 0.9805
|
| ara-
| 0.1263
|
| ara+
| 0.0471
|
We isolated the DNA from the transformed cells (with RBS, pBAD and pTet) by using the miniprep protocol and measured the concentration by [http://www.nanodrop.com/library/nd-1000-v3.7-users-manual-8.5x11.pdf NanoDrop ND-1000 Spectrophotometer].
| Sample
| Concentration (ng/µl)
|
| RBS
| 4,91
|
| pTet1
| 8,99
|
| pTet2
| 4,39
|
| pBAD1
| 3,54
|
| pBAD2
| 9,24
|
Thursday 04.07.2013
Continuation of the small-scale vesicle preparation according to the vesicle isolation protocol. Step 8-12 was completed with the exception that the pellet was resuspended in 0.5 mL DPBSS insted of 100 µL in step 11 and that the check for sterility in step 12 was not performed. The SDS-PAGE showed no signs of vesicles as viewed in figure 1.

figure 1
Monday 08.07.10
An overnight culture was started for vesicle isolation as desribed for tuesday 02.07.10.
Tuesday 09.07.2013
Today we ran PCR on 4 FP's <partinfo>Bba_K864100</partinfo>, <partinfo>Bba_K592100</partinfo>, <partinfo>Bba_E0040</partinfo>, <partinfo>Bba_E1010</partinfo> and 1 backbone <partinfo>Bba_J01101</partinfo>. We used the Phusion DNA Polymerase protocol from New England Biolabs [1] for 50 ul sample(without DMSO). All amplicons were run on a GelGreen 0.8% agarose gel. We only got a band on our RFP <partinfo>Bba_E1010</partinfo>, and it was not the expected size. We will adjust the PCR parameters and run them again tomorrow.
Step 2-11 of the small scale-vesicle preparation protocol and step 1-4 of the density gradient protocol was performed. Samples from the small-scale vesicle preparation were made ready for SDS-PAGE.
The incubation period was revised from the first attempt to isolate vesicles. The duration and other condition is summerised in the table below.
|
| Sample
| Sample
| Sample
|
| Time (h:min)
| ER2566
| MW27784 ara-
| MW27784 ara+
|
| 0:00
|
| Add 250 μL Amp (100X) and 2,5 mL of overnight culture to 250 mL of LB. Incubate at 37 C
| Add 250 μL Amp (100X) and 2,5 mL of overnight culture to 250 mL of LB. Incubate at 37 C
|
| 1:00
| Add 1 mL of overnight culture to 250 mL of LB. Incubate at 37 C
|
|
|
| 2:00
|
|
| Add 1,25 μL arabinose (1000X)
|
| 4:00
| Measure OD-600
| Measure OD-600
| Measure OD-600
|
Optical denisty (OD) at 600 nm was mesured in step 4 as indicated in the table below. No dilution of the samples was necessary.
| Sample
| OD600
|
| ER2566
| 0.1671
|
| ara-
| 0.6726
|
| ara+
| 0.5968
|
Wednesday 10.07.2013
We ran the PCR again with different parameters. Increased annealing- and elongationtime and a separate program for the backbone due to its larger size. We still only got a band for RFP <partinfo>Bba_E1010</partinfo>. In the prosess of rechecking the primers it was discovered that the primers for RFP are incorrect which explains the discrepancy in productsize. The other primers should be working so tomorrow we will redo the dilutions of the primers, increase the elongation time and lower the annealing temperature a little, add DMSO, add a bit larger templatesample and use 1-step PCR.
SDS-PAGE were run with the samples from the vesicle preparaton from the day before. There were no indication of vesicles in the samples as indicated on the gel (figure 2)
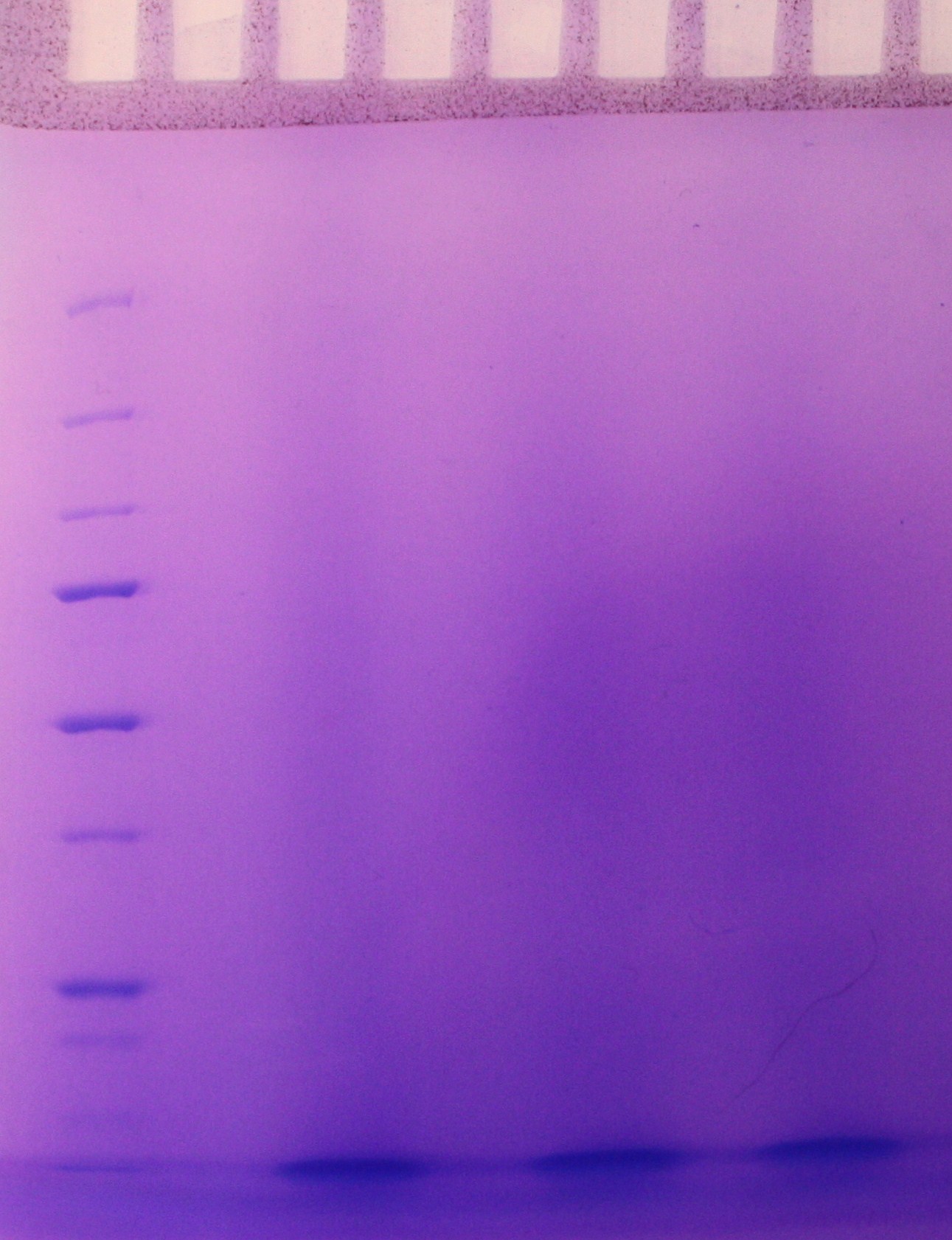
figure 2
Step 5 of the purification of vesicles by density gradient was performed. The fraction were freezed down at -80 oC.
Thursday 11.07.2013
Today we ran PCR again. This time we omitted the RFP, since we discovered that one of the primers was incorrect. We diluted the primers one more time and changed the parameters for the PCR. This time we only did one step, with 30 cycles. We also decreased the annealing temperature and increased the elongationtime. The results were more uplifting this time around after running gel electrophoreses (picture coming soon). We got a clear band on the backbone and faint bands on the GFP and SYFP. Since there was a faint band on the backbone of a larger size than the one we wanted, we added 1 µl of Dpn1 to the amplicon and put it in a 37 °C waterbath for an hour. We used the QIAGEN PCR purification kit to isolate the DNA and measured the cioncentration by [http://www.nanodrop.com/library/nd-1000-v3.7-users-manual-8.5x11.pdf NanoDrop ND-1000 Spectrophotometer].
| Sample
| Concentration (ng/µl)
|
| SYFP
| 13,16
|
| GFP
| 55,62
|
| BB
| 55,50
|
Friday 12.07.2013
We repeated the PCR on the SYFP and BFP. We changed some of the conditions (increased number of cycles and varying the template volume), but still no result.
Sunday 14.07.2013
Step 1 and 2 of the Small-scale vesicle preparation were performed. 5 mL cultures of the E.coli strains ER2566, DH5α and MW27784 were incubated in 5 mL LB at 37 oC for 8 h. 2,5 mL of these cell cultures were added to 250 mL of LB and incubated at 37 oC for 13 h.
Monday 15.07.2013
Step 3-11 of small scale-vesicle preparation protocol and step 1-4 of the density gradient protocol was performed. Samples from the small-scale vesicle preparation were made ready for SDS-PAGE.
Optical denisty (OD) at 600 nm was mesured in step 4 as indicated in the table below. The samples were diluted 1:16 with LB media.
| Sample
| OD600
|
| ER2566
| 0.133
|
| DH5α
| 0.169
|
| MW27784
| 0.198
|
A relative concentration of vesicles from the small scale preparation was mesured by adding the hydrophobic fluorecent dye [http://products.invitrogen.com/ivgn/product/T3166 FM4-64] and measuring fluorecence (RFU) (as desribed in step 12 in the small-scale vesicle preparation). Results (table below) indicated presence of vesicles in the ER2566-sample only.
| Sample
| RFU (exitation/emission at 506/750 nm)
|
| Empty
| 3
|
| Blank
| 19 814
|
| MW27784
| 19 598
|
| DH5α
| 19 254
|
| ER2566
| 40 987
|
Two rounds of PCR was run today with varying the conditions.There are still no result. For the BFP, we will wait for new primers, since the result are showing formation of primer dimers. A new transformation of the SYFP-biobrick was started to get a new template for the PCRs.
Tuesday 16.07.13
SDS-PAGE were run with the samples from the vesicle preparaton from the day before. The ER2566-sample had vesicles, where as the other samples did not contain any (figure 3)

figure 3: Ladder applied is Precision Plus ProteinTM Unstained Standards
Step 5 of the purification of vesicles by density gradient was performed. SDS-PAGE were run on the fractions from ER2566, as this was the only sample with any indication of vesicles. As indicated in figure 4, fraction 4 and 5 had clear bands, whereas the fraction 6 has pale bands. The fractions are numbered according to when whey where removed from the gradient with fraction 1 beeing the first fraction to be removed and so on.
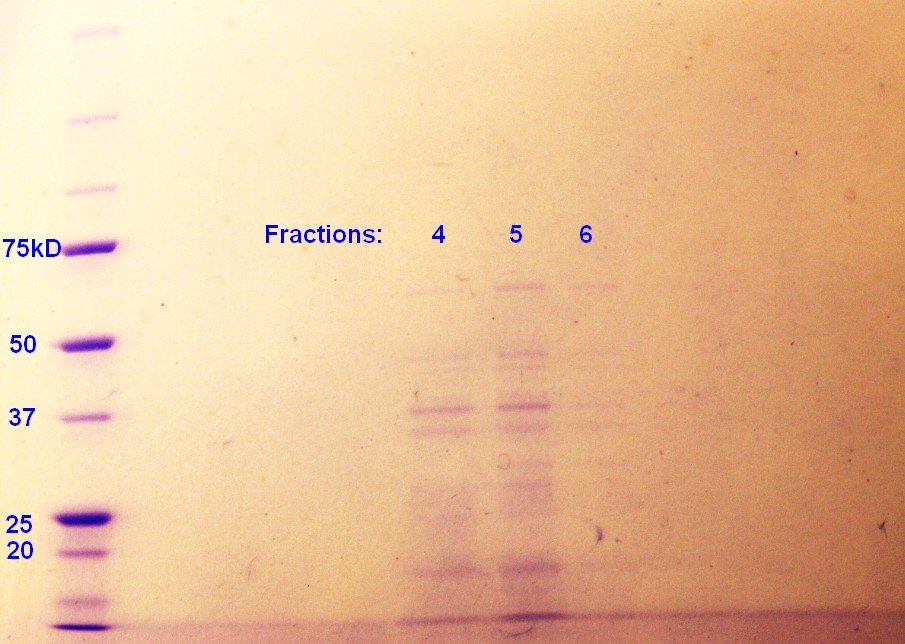
figure 4: Ladder applied is Precision Plus ProteinTM Unstained Standards
The rest of the fractions were freezed down at -80 oC. Too short incubation time in step 2 of the small-scale vesicle preparation seem to be the reason why vesicles were not isolated during the two previous attemts.
Due to postive results, the Small-Scale vesicle preparation was repeated: Step 1 and 2 of the Small-Scale vesicle preparation was performed. 5 mL cultures of the E.coli strains ER2566, DH5α and MW27784 were incubated in 5 mL LB at 37 oC for 8 h. 2.5 mL of these cell cultures were added to 250 mL of LB and incubated at 37 oC for 14 h.
Wednesday 17.07.13
Step 3-11 of small scale-vesicle preparation protocol (vesicle isolation) was performed on the ER2566, DH5α, and MW27784-samples.
Optical denisty (OD) at 600 nm was mesured in step 4 as indicated in the table below. The samples were diluted 1:10 with LB media.
| Sample
| OD600
|
| ER2566
| 0.281
|
| DH5α
| 0.293
|
| MW27784
| 0.402
|
The DH5α had no pallet after the second centrifugation, and this sample where therefore not included further in the experiement. Some of MW27784 sample was lost in step 10 of the small-scale vesicle preparation. A part of the ER2566 and MW27784-samples from the small-scale vesicle preparation were made ready for SDS-PAGE. The rest of the samples were mixed with OptiPrep (60%) to give a final OptiPrep concentration of 45% and frozen at -80oC.
A relative concentration of vesicles from the small scale preparation was mesured by adding the hydrophobic fluorecent dye [http://products.invitrogen.com/ivgn/product/T3166 FM4-64] and measuring RFU (as desribed in step 12 in the small-scale vesicle preparation). This time we also measured RFU with exitation at 515 nm and emission at 640 nm. As the last exitation and emission gave a more significant difference between the blank sample and the vesicle containing sample, this condition will be applied for later RFU-measurements. Results (table below) indicated presence of vesicles in the ER2566-sample and perhaps also in the MW27784-sample.
| Sample
| RFU (exitation/emission at 506/750 nm)
| RFU (exitation/emission at 515/640 nm)
|
| Empty
| 1
| 32
|
| Blank
| 16 536
| 1 180
|
| MW27784
| 16 335
| 1 691
|
| ER2566
| 47 191
| 44 059
|
We isolated the DNA from the transformed cells (with SYFP) by using the miniprep protocol and measured the concentration by [http://www.nanodrop.com/library/nd-1000-v3.7-users-manual-8.5x11.pdf NanoDrop ND-1000 Spectrophotometer].
| Sample
| Concentration (ng/µl)
|
| SYFP1
| 30,5
|
| SYFP2
| 20,8
|
We used the new isolated DNA as template in PCR, trying different parameters.
Thursday 18.07.13
SDS-PAGE were run with the samples from the vesicle preparaton from the day before. The ER2566-sample had a clear content of vesicles whereas the MW27784-sample had a less denser content of vesicles (figure 5). The MW27784 also seems to have a different protein profile.
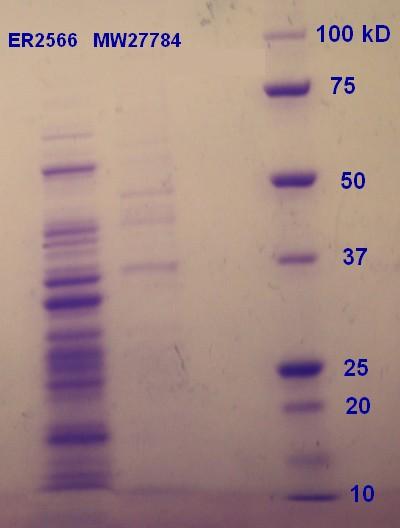
Figure 5: Ladder applied is Precision Plus ProteinTM Unstained Standards.
Friday 19.07.13
E.coli strain ER2566 was transformed with biobrick [http://parts.igem.org/Part:BBa_K530015 BBa_K530015] according to the transformation protocol. The plates were incubated for 19 hours and then moved to the fridge. The transformed ER2566 bacteria is later to be used for determination of influence of chloramphenicol on the cells and for growth duration for optimal vesicle production.
Monday 22.07.13
Three cell cultures of E.coli strain ER2566 transformed with [http://parts.igem.org/Part:BBa_K530015 BBa_K530015] was inoculated in Chl-LB and one cell culture of MW27784 were inoculated in LB for 7.5 hours at 37°C as described in step 1 of the small-scale vesicle preparation. The transformed ER2566 seems to grow slower in Chl_LB than untransformed ER2566 does in plain LB. Bigger overnight cell cultures were incubated as in step 2 of the small-scale vesicle preparation, the ER2566 cells were incolated in Chl-LB as before. The three ER2566 samples were incubated for 14, 16 and 18 hours, while the MW27784 sample was incubated for 16 hours.
Tuesday 23.07.13
Step 4-12 of the small-scale vesicle preparation were performed. In addition dilutions (10-2, 10-4 and 10-6) of the ER2566 samples were made and plating of 1/10 of the dilutions on Chl-LA was performed in step 4. The plates were left in the incubator overnight.
Optical denisty (OD) at 600 nm was mesured in step 4 as indicated in the table below. The samples were diluted 1:10 with LB media.
| Sample
| OD600
|
| ER2566 14h
| 0.247
|
| ER2566 16h
| 0.347
|
| MW27784 16h
| 0.533
|
| ER2566 18h
| 0.328
|
Samples for SDS-PAGE was prepared in step 12.
RFU was measured as decribed in step 12 of the small-scale vesicle preparation protcol. Results were as indicated in the table below:
| Sample
| RFU (exitation/emission at 515/640 nm)
|
| Empty
| 91
|
| Blank
| 6 745
|
| ER2566 14h
| 31 571
|
| ER2566 16h
| 45 089
|
| MW27784 16h
| 6 899
|
| ER2655 18h
| 22 328
|
Wednesday 24.07.13
Plates with transformed ER2566, plated the day before, were checked for colonies. As the 10-2 and 10-4 dilution had to much growth, only the 10-6-dilutions were counted for colony forming units (CFU)(table below):
| Sample
| CFU (10-6-dilution)
| CFU/mL (undiluted)
|
| ER2566 14h
| 265
| 2.65*109
|
| ER2566 16h
| 278
| 2.78*109
|
| ER2566 18h
| 273
| 2.73*109
|
It seems that transformed ER2566 reaches steady state at 14h or quickly after 14h in the 240 mL cellmedia. The CFU results corresponds well with the measured OD600 (table from the day before) except for that the OD600 of ER2566 14h is more different from the other samples compared to the CFU-results.
SDS-PAGE were run on the samples prepared the day before. There were very weak bands on the on all of the ER2566 samples (not as visible in the photograph (figure 6) as in real life) and no indication of vesicles nn the MW27784-sample.
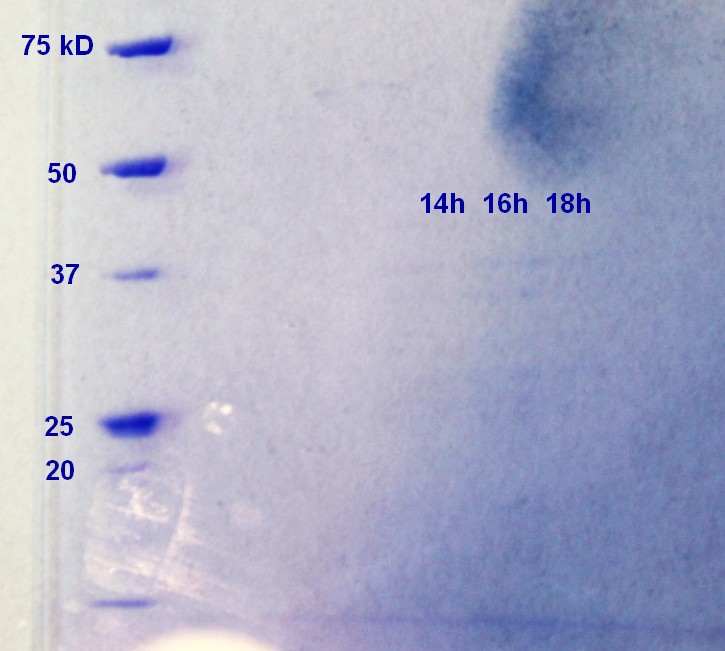
Figure 6: Ladder applied is Precision Plus ProteinTM Unstained Standards.
The results of the SDS-PAGE does not correspond well to the results of the RFU-measurements from the day before. The RFU results indicate that the concentration of vesicles is higher. We therefore started with the density gradient vesicle isolation to further confirm the same tendency. Step 1-4 of this protocol was performed.
 "
"







