26/07/13
From 2013.igem.org
(Difference between revisions)
| Line 10: | Line 10: | ||
#measured DNA concentration using nanodrop | #measured DNA concentration using nanodrop | ||
*DNA concentration (ng) | *DNA concentration (ng) | ||
| - | + | **1.1 - 131.8 | |
| - | + | **1.2 - 77.1 | |
| - | + | **1.3 - 56.8 | |
| - | + | **1.4 - 109 | |
| - | + | **1.5 - 129.1 | |
| - | + | **1.2 - 93.3 | |
#Restriction enzyme digest of the DNA samples | #Restriction enzyme digest of the DNA samples | ||
*enzymes: XbaI, PstI | *enzymes: XbaI, PstI | ||
*master mix: | *master mix: | ||
| - | Buffer - 14ul | + | **Buffer - 14ul |
| - | H2O - 88.2ul | + | **H2O - 88.2ul |
| - | XbaI - 1.4ul | + | **XbaI - 1.4ul |
| - | PstI - 1.4ul | + | **PstI - 1.4ul |
| - | Total volume - 105ul | + | **Total volume - 105ul |
| - | Each sample contained 5ul of DNA and 15ul of the master mix. | + | *Each sample contained 5ul of DNA and 15ul of the master mix. |
| - | The total amount of DNA in each sample was 200ng | + | *The total amount of DNA in each sample was 200ng |
| - | To calculate it we divided 200ng by the concentration of each sample | + | *To calculate it we divided 200ng by the concentration of each sample |
| - | The rest of the volume was made up with water to give a total of 5ul. | + | *The rest of the volume was made up with water to give a total of 5ul. |
| - | The samples were them incubated in a water bath at 37C by an hour | + | *The samples were them incubated in a water bath at 37C by an hour |
| - | + | #Agorose Gel preparation | |
| - | + | **20ml of 5xTBE | |
| - | + | **80ml of water | |
| - | + | **0.8g of agarose | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | *Boil for 3 minutes, 1 minute at a time. | ||
| + | *Let it cool and add 5ul of ethidium bromide. | ||
| + | *Pour and let set for 30 mins. | ||
*Adding dye and and marker | *Adding dye and and marker | ||
| - | Used orange g loading dye. | + | *Used orange g loading dye. |
| - | + | **2ul to each sample | |
| - | Used 1kb ladder, 5ul per 1 marker well. | + | *Used 1kb ladder, 5ul per 1 marker well. |
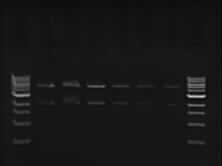
| - | + | #Run the gel | |
Revision as of 09:38, 29 July 2013
- purification of the limonene biobrick - BBa_24
- Plasmid Volume
-1.1 - 40ul -1.2 - 40ul -1.3 - 46ul -1.4 - 39ul -1.5 - 41ul -1.2 - 32ul
- measured DNA concentration using nanodrop
- DNA concentration (ng)
- 1.1 - 131.8
- 1.2 - 77.1
- 1.3 - 56.8
- 1.4 - 109
- 1.5 - 129.1
- 1.2 - 93.3
- Restriction enzyme digest of the DNA samples
- enzymes: XbaI, PstI
- master mix:
- Buffer - 14ul
- H2O - 88.2ul
- XbaI - 1.4ul
- PstI - 1.4ul
- Total volume - 105ul
- Each sample contained 5ul of DNA and 15ul of the master mix.
- The total amount of DNA in each sample was 200ng
- To calculate it we divided 200ng by the concentration of each sample
- The rest of the volume was made up with water to give a total of 5ul.
- The samples were them incubated in a water bath at 37C by an hour
- Agorose Gel preparation
- 20ml of 5xTBE
- 80ml of water
- 0.8g of agarose
- Boil for 3 minutes, 1 minute at a time.
- Let it cool and add 5ul of ethidium bromide.
- Pour and let set for 30 mins.
- Adding dye and and marker
- Used orange g loading dye.
- 2ul to each sample
- Used 1kb ladder, 5ul per 1 marker well.
- Run the gel
 "
"