Team:Paris Saclay/Notebook/August/9
From 2013.igem.org
(Difference between revisions)
| Line 18: | Line 18: | ||
| style="width:350px;border:1px solid black;" | [[File:Ps908transduction.jpg|350px]] | | style="width:350px;border:1px solid black;" | [[File:Ps908transduction.jpg|350px]] | ||
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
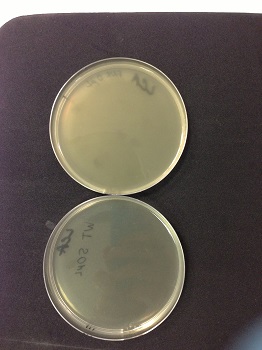
| - | ''' | + | '''Picture: lysed cells comparison.''' |
| - | * 0µl phage(control): the petri dish is cloudy, bacteria are not lysed | + | * 0µl phage(control): the petri dish is cloudy, bacteria are not lysed. |
* 50µl phage: the petri dish is clear, bacteria are lysed by phages. | * 50µl phage: the petri dish is clear, bacteria are lysed by phages. | ||
*We used a wild type strain to keep a stock and a Δ ''fnr E. coli'' strain to obtain phages that might have encapsidated a Δfnr::Km fragment. | *We used a wild type strain to keep a stock and a Δ ''fnr E. coli'' strain to obtain phages that might have encapsidated a Δfnr::Km fragment. | ||
|} | |} | ||
| - | + | 10µl,50µl and 100µl petri dishes are clear so phages are multiplied. | |
| + | |||
| + | We let the antibiotic over night to select the right strain. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{Team:Paris_Saclay/incl_fin}} | {{Team:Paris_Saclay/incl_fin}} | ||
Revision as of 12:26, 11 August 2013
Contents |
Notebook : August 9
Lab work
A - Aerobic/Anaerobic regulation system
Obtaining Δ fnr E. coli strain by transduction to test our biobricks
Abdou, Anais, Damir, Nadia, XiaoJing
We do the exprience again because the lysis made on wensday didn't happen. It's probably because the phage strain was too old ( 2001)
Protocol : transduction
10µl,50µl and 100µl petri dishes are clear so phages are multiplied.
We let the antibiotic over night to select the right strain.
 "
"