Team:DTU-Denmark/Notebook/19 July 2013
From 2013.igem.org
(→Conclusion) |
(→Conclusion) |
||
| Line 112: | Line 112: | ||
*No bands for cycAX in pZA21 | *No bands for cycAX in pZA21 | ||
*Two bands are present in colony 3 and 4 of RFP in pZA21 and one in colony 20. | *Two bands are present in colony 3 and 4 of RFP in pZA21 and one in colony 20. | ||
| - | + | ||
Navigate to the [[Team:DTU-Denmark/Notebook/18_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/22_July_2013|Next]] Entry | Navigate to the [[Team:DTU-Denmark/Notebook/18_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/22_July_2013|Next]] Entry | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Revision as of 12:14, 23 August 2013
19 July 2013
Navigate to the Previous or the Next Entry
Contents |
208
Main purpose
- plasmid isolation
- colony PCR to verify transformants from 18-07-2013.
- gel electrophoresis to verify insert after colony PCR (AMO, HAO), plasmid isolation
- gel electrophoresis to verify plasmid isolation
- restriction analysis of isolated plasmids
- gel electrophoresis to verify restriction analysis
Who was in the lab
Gosia, Henrike
Procedure
Plasmid isolation
According to protocol in QIAprep Spin Miniprep Kit
Colony PCR
Master mix was prepared based on standard colony PCR procedure.
For AMO primers 29a, 29b were used (no USER primers this time) and for HAO primers 28a, 28b were used. We used Phusion polymerase. We obtained 7 transformants after yesterday transformation with AMO in pZA21 and 3 transformants which may contain HAO in pZA21. On negative control plate 3 colonies grew.
PCR parameters according to standard reaction with differences in annealing temperatures (for AMO -> 54°C, for HAO -> 56°C) and with 3 min of extension time. Expected fragments lengths are AMO - 3074 bp, HAO - 2816 bp.
Restriction analysis
Restriction analysis was performed with EcoRI enzyme.
| Compound | Volume uL |
|---|---|
| DNA (miniprep) | 10 |
| Buffer for EcoRI 10x | 2 |
| water | 7,5 |
| EcoRI enzyme | 0,5 |
Incubation for 1-2 hours at 37°C.
For plasmid pZA21 with cycAX we expect 4 DNA fragments after digestion with EcoRI, lengths are as follows: 2283 bp, 787 bp, 710 bp, 150 bp. If insert will not be present only two fragments (probably only one visible) - 70 bp, 2350 bp.
For plasmid pZA21 with RFP we expect 2 DNA fragments after digestion with EcoRI, lengths are as follows: 2250 bp and 750 bp. If insert will not be present only two fragments (probably only one visible) - 70 bp, 2350 bp.
Results
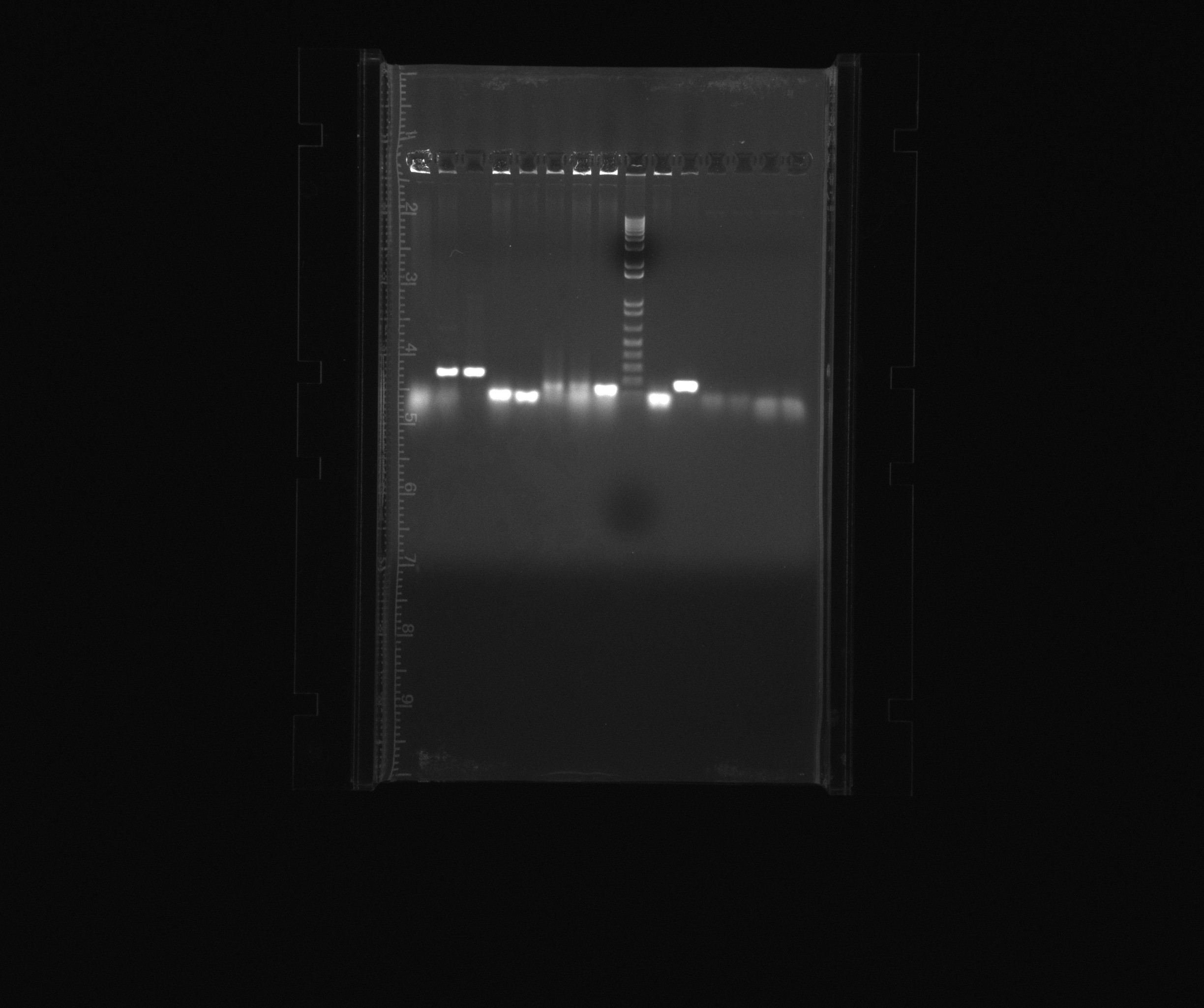
1% gel of results of today's colony PCR and yesterday's PCR to add his-tags
- 1: HAO, colony PCR
- 2: HAO, colony PCR
- 3: HAO, colony PCR
- 4: AMO, colony PCR
- 5: AMO, colony PCR
- 6: AMO, colony PCR
- 7: AMO, colony PCR
- 8: AMO, colony PCR
- 9: 1kb ladder
- 10: AMO, colony PCR
- 11: AMO, colony PCR
- 12: TAT construct, his-tag, program ramp1
- 12: TAT construct, his-tag, program ramp2
- 12: Sec construct, his-tag, program ramp1
- 12: Sec construct, his-tag, program ramp2
1% gel of plasmids isolated by qiagen spin miniprep kit
- 1: 1kb ladder
- 2: cycAX from isolation of the 19-07
- 3: cycAX from isolation of the 19-07 dub
- 4: cycAX from isolation of the 17-07
- 5: RFP from isolation on the 19-07
- 6: RFP from isolation on the 19-07 dub
- 7: RFP from isolation on the 19-07 trip
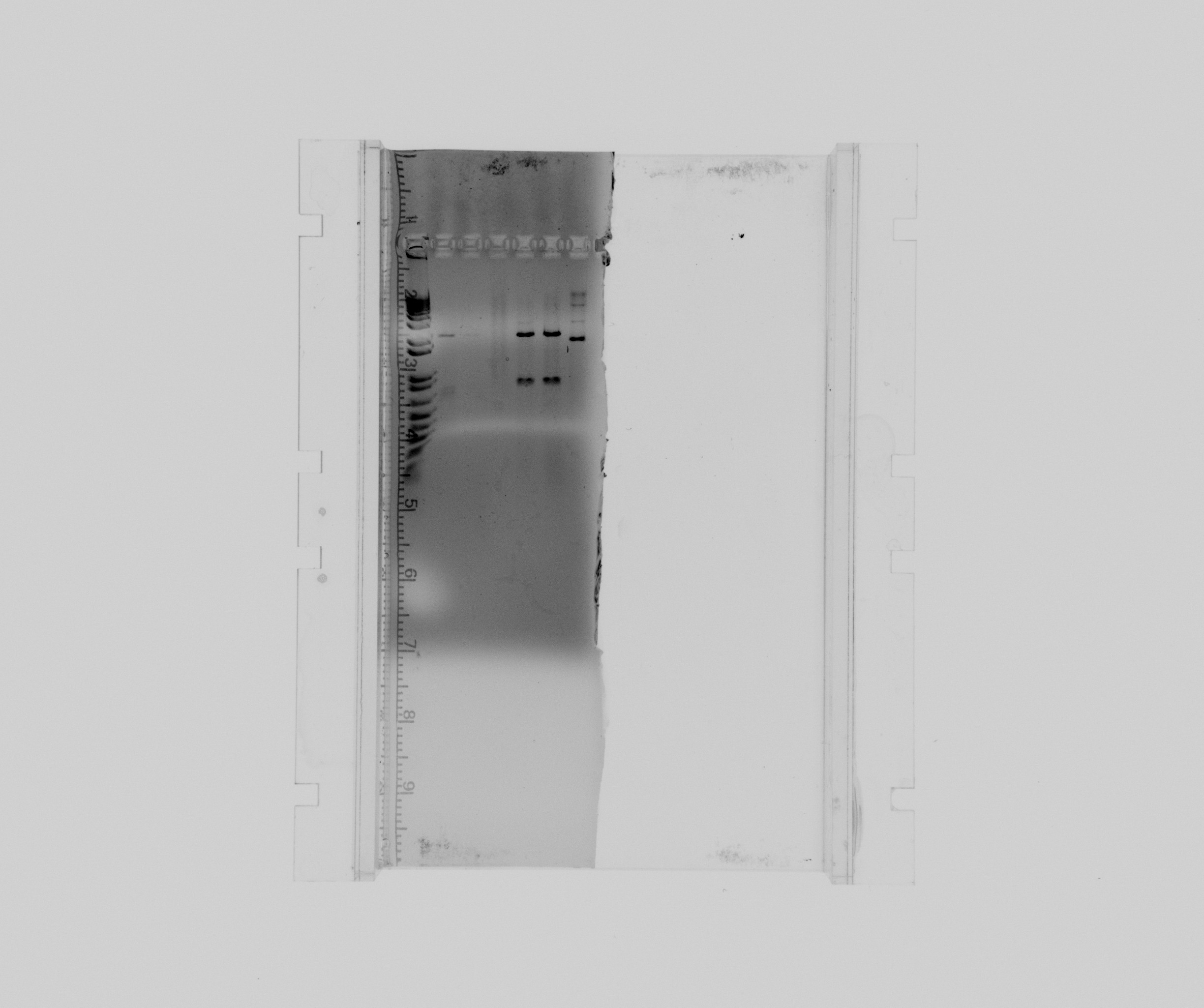
1% gel showing result of restriction analysis
- 1 - 1kb ladder
- 2 - cycAX in pZA21 after EcoRI digestion, sample 1, colony 12
- 3 - cycAX in pZA21 after EcoRI digestion, sample 2, colony 12
- 4 - cycAX in pZA21 after EcoRI digestion, sample 3, colony 12
- 5 - RFP in pZA21 after EcoRI digestion, colony 3
- 6 - RFP in pZA21 after EcoRI digestion, colony 4
- 7 - RFP in pZA21 after EcoRI digestion, colony 20
Conclusion
After ExoRI digestion:
- No bands for cycAX in pZA21
- Two bands are present in colony 3 and 4 of RFP in pZA21 and one in colony 20.
Navigate to the Previous or the Next Entry
 "
"