From 2013.igem.org
Sep 3
File:Igku Notebook Sep3 1.jpg
File:Igku Notebook Sep3 2.jpg
File:Igku Notebook Sep3 3 1.jpg
File:Igku Notebook Sep3 3 2.jpg
File:Igku Notebook Sep3 4 1.jpg
File:Igku Notebook Sep3 4 2.jpg
File:Igku Notebook Sep3 4 3.jpg
File:Igku Notebook Sep3 4 4.jpg
File:Igku Notebook Sep3 5 1.jpg
File:Igku Notebook Sep3 5 2.jpg
File:Igku Notebook Sep3 5 3.jpg
File:Igku Notebook Sep3 5 4.jpg
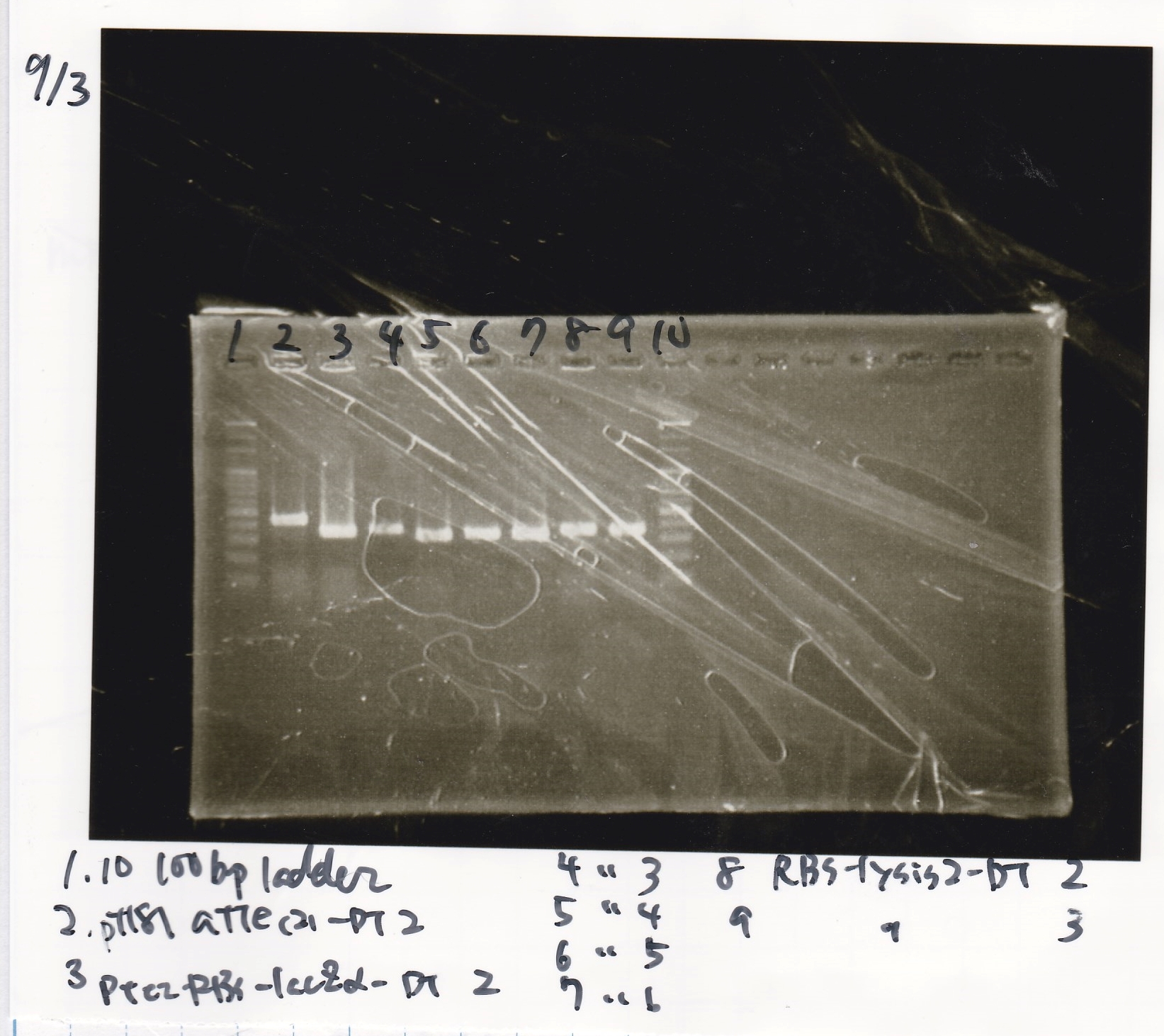
Electrophoresis
Kozima
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | pT181 attenuater(2)-DT2(738)
|
| 3 | Ptet-RBS-lacZα-DT-2(756)
|
| 4 | Ptet-RBS-lacZα-DT-3
|
| 5 | Ptet-RBS-lacZα-DT-4
|
| 6 | Ptet-RBS-lacZα-DT-5
|
| 7 | Ptet-RBS-lacZα-DT-6
|
| 8 | RBS-lysis2-DT-2(985)
|
| 9 | RBS-lysis2-DT-3
|
| 10 | 100bp ladder
|
Miniprep
Tatsui
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/2 14:00 pT181 attenuator | 418.4 | 1.82 | 1.80
|
| 9/2 14:00 pT181 antisense | 362.2 | 1.92 | 1.96
|
| 9/2 14:00 Spinach | 438.3 | 1.86 | 1.99
|
| 9/2 14:00 tetR-apt 12_1R | 567.5 | 1.90 | 1.99
|
| 9/2 21:00 pT181 attenuater | 320.9 | 1.90 | 2.15
|
| 9/2 21:00 Spinach | 457.0 | 1.88 | 2.24
|
Liquid Culture
Kozima
| Sample | medium
|
| Spinach-DT(9/2 master plate) | plusgrow(CP)4ml
|
Restriction Enzyme Digestion
Kozima
| | DNA | EcoR1 | Spe1 | Xba1 | Pst1 | 10xBuffer B(EcoR1&Spe1) | 10xBuffer D(Xba1&Pst1) | 100xBSA | MilliQ | total
|
| 9/3 pT181attenuater 14:00 418.4ng/µL | 4.8µL | 1.0µL | 1.0µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 19.9µL | 30µL
|
| 9/3 pT181attenuater 14:00 418.4ng/µL | 0.2µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.7µL | 10µL
|
| 9/3 pT181attenuater 14:00 418.4ng/µL | 4.8µL | 0µL | 0µL | 1.0µL | 1.0µL | 0µL | 3µL | 0.3µL | 19.9µL | 30µL
|
| 9/3 pT181attenuater 14:00 418.4ng/µL | 0.2µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.7µL | 10µL
|
| | DNA | EcoR1 | Spe1 | Xba1 | Pst1 | 10xBuffer B(EcoR1&Spe1) | 10xBuffer D(Xba1&Pst1) | 100xBSA | MilliQ | total
|
| 9/3 pT181antisense 14:00 362.2ng/µL | 5.5µL | 1.0µL | 1.0µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 19.2µL | 30µL
|
| 9/3 pT181attenuater 14:00 418.4ng/µL | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.6µL | 10µL
|
| 9/3 pT181attenuater 14:00 418.4ng/µL | 5.5µL | 0µL | 0µL | 1.0µL | 1.0µL | 0µL | 3µL | 0.3µL | 19.2µL | 30µL
|
| 9/3 pT181attenuater 14:00 418.4ng/µL | 0.3µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.6µL | 10µL
|
| | DNA | EcoR1 | Spe1 | 10xBuffer B | 100xBSA | MilliQ | total
|
| 9/3 apt 12_1R 14:00 507.5ng/µL | 3.9µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 20.8µL | 30µL
|
| 9/3 apt 12_1R 14:00 507.5ng/µL | 0.2µL | 0µL | 0µL | 1µL | 0.1µL | 8.7µL | 10µL
|
| | DNA | EcoR1 | Xba1 | 10xBuffer D | 100xBSA | MilliQ | total
|
| 8/29 DT 166.2ng/µL | 12µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 12.7µL | 30µL
|
| 8/29 DT 166.2ng/µL | 0.6µL | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL
|
| | DNA | EcoR1 | Spe1 | 10xBuffer B | 100xBSA | MilliQ | total
|
| 8/22 RBS-lysis1(2) 96ng/µL | 21µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 3.7µL | 30µL
|
| 8/22 RBS-lysis1(2) 96ng/µL | 1.0µL | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
| | DNA | EcoR1 | Spe1 | 10xBuffer B | 100xBSA | MilliQ | total
|
| 8/30 RBS-lysis2 220ng/µL | 9.1µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 15.6µL | 30µL
|
| 8/30 RBS-lysis2 220ng/µL | 0.5µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
| | DNA | EcoR1 | Spe1 | 10xBuffer B | 100xBSA | MilliQ | total
|
| 8/20 RBS-lysis3(1) 282ng/µL | 7.1µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 17.6µL | 30µL
|
| 8/20 RBS-lysis3(1) 282ng/µL | 0.4µL | 0µL | 0µL | 1µL | 0.1µL | 8.5µL | 10µL
|
PCR
Hirano
| Plac(1) plasmid 153.7ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer (KaiA_BsaI_Mut_fwd) | primer (KaiA_BsaI_Mut_rer | KOD plus-ver.2 | MilliQ | total
|
| 0.13 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16.37 | 25
|
| KaiB (plasmid) 20.2ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer (GG2_RBS_KaiB_fwd) | primer (KaiB_GG3_rer | KOD plus-ver.2 | MilliQ | total
|
| 1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| °C | °C | °C | °C | --
|
| (◎Д◎) | | | |
|
Mutatation PCR
Colony PCR
Restriction Enzyme Digestion
Liquid Culture
 "
"
