Template:Kyoto/Notebook/Sep 27
From 2013.igem.org
Sep 27
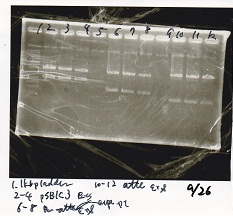
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kbp ladder | -- |
| 2 | pSB1C3 | EcoRI+SpeI |
| 3 | ||
| 4 | ||
| 6 | Pcon-attenuator-aptmer-DT | EcoRI+SpeI |
| 7 | ||
| 8 | ||
| 10 | Pcon-attenuator-aptmer-DT | EcoRI+SpeI |
| 11 | ||
| 12 |
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-attenuator-DT(EcoRI+SpeI) | 6.8 | 1.98 | 0.43 |
| pT181-attenuator(EcoRI+SpeI) | 7.5 | 1.84 | 0.06 |
| pSB1C3(EcoRI+SpeI) | 17.7 | 1.97 | 0.71 |
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kbp ladder | -- |
| 3 | pSB4K5 | EcoRI+PstI |
| 4 | pSB4K5 | EcoRI+PstI |
| 5 | pSB4K5 | EcoRI+PstI |
| 7 | pSB6A1 | EcoRI+PstI |
| 8 | pSB6A1 | EcoRI+PstI |
| 9 | pSB6A1 | EcoRI+PstI |
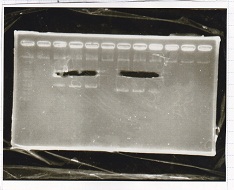
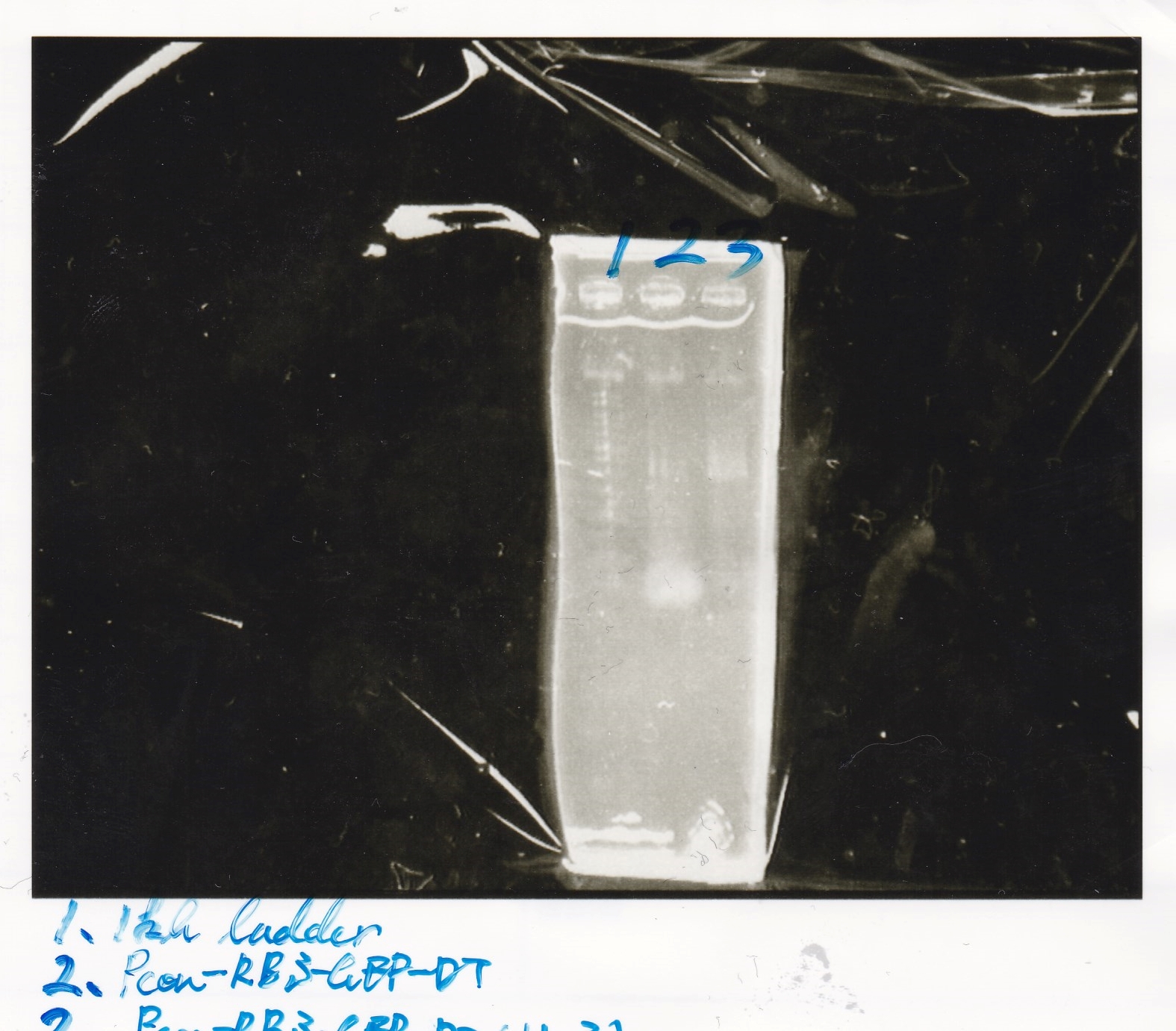
Electrophoresis
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | pSB4K5 | EcoRI | PstI |
| 2 | pSB4K5 | - | - |
| 3 | 1kbp ladder | - | - |
| 4 | Pcon-attenuator | EcoRI | SpeI |
| 5 | Pcon-attenuator | - | - |
| Lane | Sample | Enzyme1 | Enzyme2 |
|---|---|---|---|
| 1 | RBS-GFP-DT | XbaI | PstI |
| 2 | RBS-GFP-DT | - | - |
| 3 | RBS-GFP-DT | EcoRI | XbaI |
| 4 | 1kbp ladder | - | - |
| 5 | Ptet | EcoRI | SpeI |
| 6 | Ptet | - | - |
Liquid Culture
| Sample | medium |
|---|---|
| Pcon-GFP | Amp |
| RBS-GFP-DT | CP |
| Pcon-attenuator | Amp |
| Pcon-attenuator-aptamer-DT | Amp |
| Pcon-tetR-DT | Amp |
Ligation
| state | Vector | Inserter | Ligation High ver.2 | ||
|---|---|---|---|---|---|
| experiment | 9/17 pSB1C3 (EcoRI+SpeI) 10.0ng/µL | 10µL | 9/26 pT181 attenuator(EcoRI+SpeI) 7.5ng/µL | 1.3µL | 3.5 µL |
| experiment | 9/25 DT (EcoRI+XbaI) 42.8ng/µL | 2.3µL | 9/22 Pcon-pT181 antisense(EcoRI+SpeI) 46.7ng/µL | 1.5µL | 1.9 µL |
| experiment | 9/27 pSB4K5(EcoRI+PstI) 12.9ng/µL | 7.8µL | 9/10 Pcon-attenuator(EcoRI+SpeI) 13.8ng/µL | 8.0µL | 3.5 µL |
| experiment | 9/27 pSB4K5(EcoRI+PstI) 12.9ng/µL | 7.8µL | 9/25 RBS-GFP-DT(XbaI+PstI) 13.9ng/µL | 12µL | 3.5 µL |
| experiment | 9/8 pSB4K5 (EcoRI+SpeI) 17.6ng/µL | xx µL | 8/21 Pcon-GFP-DT(EcoRI+SpeI) 11ng/µL | 12.6µL | xx µL |
Liquid Culture
| Sample | medium |
|---|---|
| Pcon-spinach-DT | plusgrow Amp |
| Pcon-tetRaptamer-DT | plusgrow Amp |
| Pcon-antisense-spinach-DT | plusgrow Amp |
| spinach-DT | plusgrow CP |
RNA Extraction
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| 9/26 Pcon-GFP-DT | 423.0 | 1.83 | 1.46 |
| 9/26 Pcon-GFP-DT K | 298.7 | 1.82 | 1.50 |
Electrophoresis
Miniprep
| DNA |
|---|
| Pcon-aptamer-DT |
| Pcon-spinach-DT |
| Pcon-antisense |
| Pcon-tetR-DT |
| Pcon-attenuator-DT |
| Pcon-antisense-spinach-DT |
Transformation
| Name1 | Name2 | Sample1(µL) | Sample2(µL) | Competent Cells(µL) | Total(µL) |
|---|---|---|---|---|---|
| Pcon-attenuator+RBS-GFP-DT(4K5) | Pcon-attenuator-DT(1A2) | 2 | 1 | 30 | 33 |
| Pcon-attenuator+RBS-GFP-DT(4K5) | Pcon-antisense-spinach-DT(1A2) | 2 | 1 | 30 | 33 |
| Pcon-attenuator+RBS-GFP-DT(4K5) | Pcon-tetR aptamer-DT(1A2) | 2 | 1 | 30 | 33 |
| Pcon-RBS-GFP-DT(4K5) | Pcon-attenuator-DT(1A2) | 2 | 1 | 30 | 33 |
| Pcon-RBS-GFP-DT(4K5) | Pcon-antisense-spinach-DT(1A2) | 2 | 1 | 30 | 33 |
| Pcon-RBS-GFP-DT(4K5) | Pcon-tetR aptamer-DT(1A2) | 2 | 1 | 30 | 33 |
| Pcon-attenuator+RBS-GFP-DT(4K5) | -- | 2 | -- | 20 | 22 |
| Pcon-RBS-GFP-DT(4K5) | -- | 2 | -- | 20 | 22 |
| attenuator(1C3) | -- | 2 | -- | 20 | 22 |
Miniprep
| DNA | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| Pcon-RBS-GFP-DT | 137.6 | 1.95 | 1.78 |
| RBS-GFP-DT | 77.0 | 2.06 | 1.96 |
| 231.0 | 1.89 | 1.89 | |
| Pcon-antisense-Spinach-DT(2) | 130.0 | 1.56 | 1.62 |
| Pcon-aptamer-DT | 218.8 | 1.92 | 1.72 |
| Pcon-attenuator-DT | 180.7 | 1.95 | 1.78 |
| Pcon-Spinach-DT | 177.6 | 1.96 | 1.06 |
| Pcon-antisense-Spinach-DT(1) | 181.1 | 1.96 | 1.92 |
| Pcon-antisense | 152.9 | 2.02 | 1.96 |
| Pcon-RBS+ezR-DT | 164.1 | 1.93 | 1.91 |
Restriction Enzyme Digestion
| 9/27 RBS-GFP-DT | XbaI | PstI | Buffer | BSA | MilliQ | total | |
|---|---|---|---|---|---|---|---|
| 2 cuts | 12µL | 1µL | 1µL | 3µL | 3µL | 20µL | 40µL |
| NC | 1.3µL | 0µL | 0µL | 1µL | 1µL | 6.7µL | 10µL |
| 9/27 Pcon-tetR-DT | EcoRI | PstI | Buffer | MilliQ | total | |
|---|---|---|---|---|---|---|
| 2 cuts | 12.2µL | 1µL | 1µL | 3µL | 12.8µL | 30µL |
| NC | 0.6µL | 0µL | 0µL | 1µL | 8.4µL | 10µL |
RNA Extraction
| Name | concentration[µg/mL] | 260/280 | 260/230 |
|---|---|---|---|
| 9/27 Pcon-RBS-GFP-DT | 686.6 | 1.99 | 1.34 |
| 9/27 Pcon-RBS-GFP-DT (+High salt) | 127.6 | 1.76 | 0.27 |
DNA sythesis
| genome DNA | DNA volume | genome DNA wipeout buffer | RNase freewater | RT | RT buffer | Primer Mix | total |
|---|---|---|---|---|---|---|---|
| 9/27 Pcon-RBS-GFP-DT | 0.8µL | 1µL | 5.2µL | 0.5µL | 2µL | 0.5µL | 10µL |
| 9/27 pcon-RBS-GFP-DT(+high salt) | 2.5µL | 1µL | 3.5µL | 0.5µL | 2µL | 0.5µL | 10µL |
| 9/27 Pcon-RBS-GFP-DT(revised concentration) | 0.5µL | 1µL | 5.5µL | 0.5µL | 2µL | 0.5µL | 10µL |
Electrophoresis
Liquid Culture
| Sample | medium |
|---|---|
| Pcon spinach DT | plusgrow 2µL(Amp) |
| Pcon aptamer DT | plusgrow 2µL(Amp) |
| Pcon antisense Spinach DT | plusgrow 2µL(Amp) |
| Spinach DT | plusgrow 4µL(CP) |
RT PCR
| genome DNA | KOD plus | 10x buffer for KOD-plus-ver.2 | 2nM dNTPs | 25mM MgSO4 | F primer | R primer | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.5 | 1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 49.8°C | 68°C | -- |
| 2min | 10sec | 30sec | 10sec |
Electrophoresis
| Lane | Sample |
|---|---|
| 1 | Pcon-GFP-DT(GFP) |
| 2 | Pcon-GFP-DT(internal) |
| 3 | Pcon-GFP-DT(HS)(GFP) |
| 4 | Pcon-GFP-DT(internal) |
| Lane | Sample |
|---|---|
| 1 | Pcon-GFP-DT(GFP) |
| 2 | Pcon-GFP-DT(internal) |
| 3 | 100bp ladder |
Digesting cut check
| Lane | Sample |
|---|---|
| 1 | RBS-GFP-DT(XbaI&PstI) |
| 2 | RBS-GFP-DT NC(XbaI&PstI) |
| 3 | 1kbp ladder |
| 4 | Pcon-RBS-tetR-DT(EcoRI&SpeI) |
| 5 | Pcon-RBS-tetR-DT NC(EcoRI&SpeI) |
Gel Extraction
| Lane | DNA | Enzyme |
|---|---|---|
| 1 | 1kbp ladder | -- |
| 3 | RBS-GFP-DT | Xbal+PstI |
| 4 | RBS-GFP-DT | Xbal+PstI |
| 5 | RBS-GFP-DT | Xbal+PstI |
| 7 | 1kbp ladder | -- |
| 9 | Pcon-tetR-DT | EcoRI+SpeI |
| 10 | Pcon-tetR-DT | EcoRI+SpeI |
| 11 | Pcon-tetR-DT | EcoRI+SpeI |
 "
"