Team:DTU-Denmark/Notebook/11 July 2013
From 2013.igem.org
Contents |
208
Main purpose
- PCR with USER primers to combine Nir operon with pZA21
- PCR with USER primer to test the activity of the promoter AraB.
Who was in the lab
Ariadni,Henrike,Julia,Jakob, Gosia
Procedure
Combine Nir operon with pZA21
Following Mathildes Protocol for DpnI digestion of our linear pZA21 w. Uracil incerts
Mastermix for 7 PCR tubes, Using the mastermix protocol we got from Mathilde.
triplets of each of the two Nir Operon extracts that we have made yesterday, including one negative control:
- Nir 1(a, b, c)
- Nir 2(a, b, c)
Using the following PCR program:
| time | Temp C | |
|---|---|---|
| Denaturing | 2 min | 98 |
| start of loop | 10 sec | 98 |
| annealing | 30 sec | 60 |
| elongation | 9 min | 72 |
| goto start of loop 35 times | ||
| hold | 10 |
PCR for promoter test
We want to test the inducible AraBAD promoter using plasmid pZA21 without its native promoter and RFP gene. We performed 3 different PCR reactions using user primers. Samples are named:
- 1,2,3 -> pZA21 with no native promoter - Primers 13a and 13b, template pZA21 miniprep
- 4,5,6 -> RFP - Primers 14a and 14b, template RFP in pZE21 miniprep
- 7,8,9 -> AraBAD promoter - Primers 12a and 12b, template biobrick K808000
Settings for PCR of 1,2,3
| Temperature (oC) | Time (min) | Rounds |
|---|---|---|
| 98 | 2:00 | 1 |
| 98 | 0:20 | 35 |
| 58 | 0:45 | 35 |
| 72 | 2:00 | 35 |
| 72 | 5:00 | 1 |
| 10 | ∞ | - |
Settings for PCR of 4-9
| Temperature (oC) | Time (min) | Rounds |
|---|---|---|
| 98 | 2:00 | 1 |
| 98 | 0:20 | 35 |
| 61 | 0:45 | 35 |
| 72 | 1:00 | 35 |
| 72 | 5:00 | 1 |
| 10 | ∞ | - |
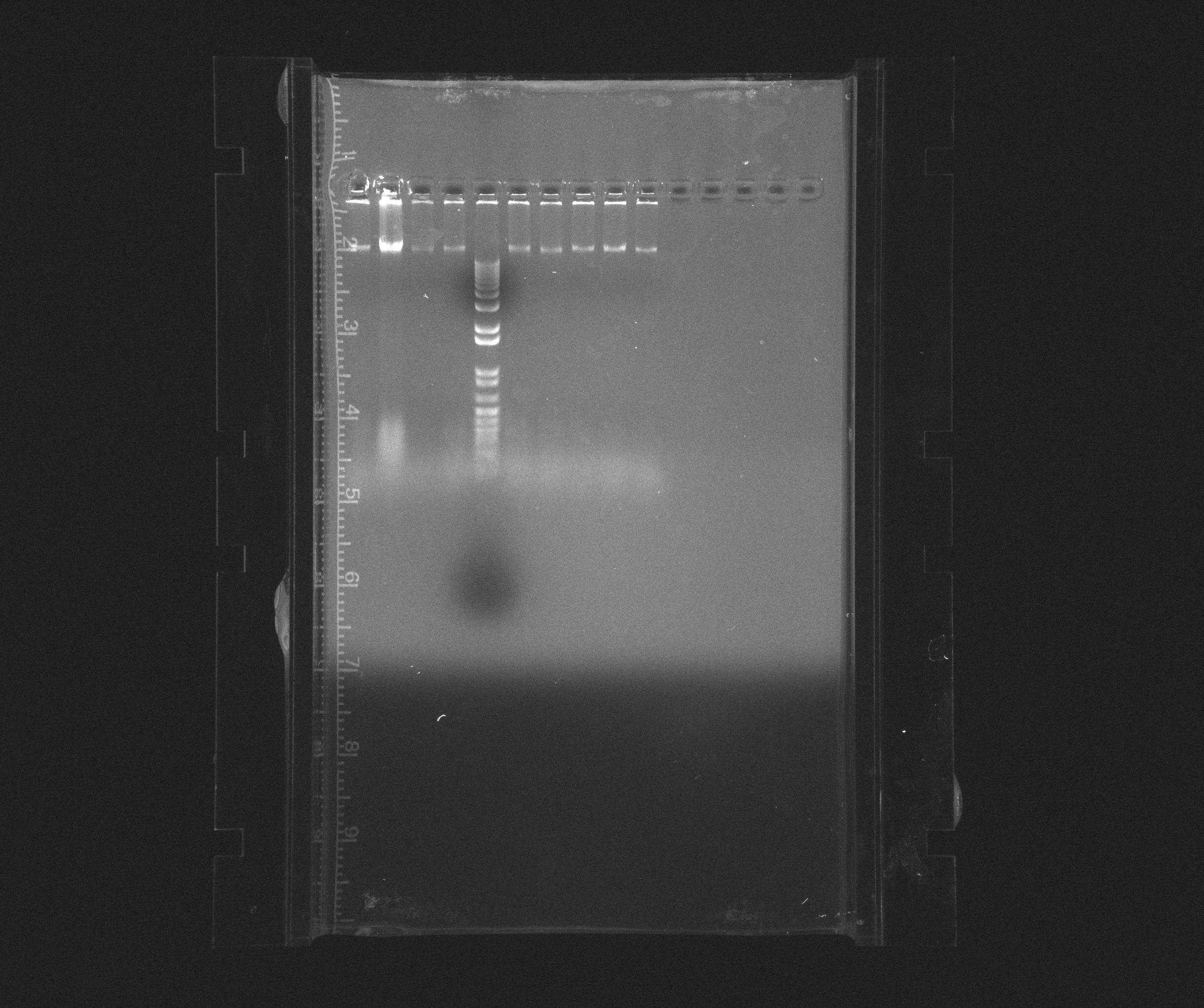
Gel analyses of Nir operon, PCR product from 10-07-2013
We run 0,8% agarose gel with 9 samples.
Result
0.8 % Agorase Gel (Nir operon)
Wells
- 1: Nir extension time 2:00
- 2: Nir extension time 2:00
- 3: Nir extension time 2:00
- 4: Nir extension time 3:00
- 5: 1kb ladder
- 6: Nir extension time 3:00
- 7: Nir extension time 3:00
- 8: Nir extension time 4:00
- 9: Nir extension time 4:00
- 10: Nir extension time 4:00
 "
"