Team:Exeter/Modelling
From 2013.igem.org
Modelling
Abstract
We have developed a stochastic model of a colour bio-camera system in E.coli. The model takes the light incident of the bacteria and stochastically estimates the light regulated expression of pigment proteins over time. The population of proteins over time is combined with their absorption spectra and plotted as a surface showing the cell’s absorption spectrum over time. There have been insufficient experimental results to test the model with rates and absorption spectra taken from literature and theoretical conjecture. This prevents us from drawing accurate conclusions. However our preliminary results suggest that a collection of cells will show a reliable colour but with a very high variance between individuals. This reduces the spatial control of expression and thus the resolution of any bio-camera. One way to counter this problem and the problem of one pathway dominating the others is to insert multiple copies of the light regulated genes. This causes spatial control to be increased as the behaviour of each individual is more predictable. This model requires experimental data to test it and update it to produce an accurate description of our biological system. In its current for it provides a well documented framework for future rule-based modelling of similar systems.
Introduction
Our model predicts how the absorption spectrum of a modified E. coli changes over time in response to a given incident light spectrum. It can estimate key properties of the system such as stability, development time of the image, and the balance of colours.
The model is split into three sections. Initially, the reaction of each sensor species to a given incident light spectrum is calculated. This information feeds to the heart of the model, which involves simulating the biochemistry of our designed pathways. Finally the data from the simulation is used to plot the absorption spectrum of the system as it varies with time.
Currently the model is based on results from literature and theoretical conjecture as sufficient experimental results are not available. The motivation for the model is to numerically characterize our bio-bricks for future use and to help us create the first colour coliroid. With experimental results the model could readily be updated to fulfill its original mandate.
Control of bacteria using light has many applications. Di-chromatic control of bacteria has been achieved, the challenge now is for Tri-chromatic control. Our lab project aims to produce a full colour ‘coliroid’ to demonstrate tri-chromatic control of bacteria. The modified E.coli are designed to carry three independent light sensitive pathways that each control the production of a specific pigment protein.
A reliable computer model is essential for accurate implementation of tri-chromatic control. In the case of the theoretical bio-camera it is required to achieve an even balance of colours and optimum development time. In other applications it is essential to have accurate temporal control over the expression of light regulated genes.
The model is based around KaSim, a stochastic simulator that reads Kappa, a rule based language. Stochastic simulation has the advantage over deterministic simulation because it replicates the random nature of brownian motion in cells more accurately. Rule based languages are particularly useful for biochemical signalling as the physical reactions are readily converted into rules. Matlab is used to process the signal from the incident light to the KaSim simulation. The outputs of the simulation is used in MATLAB to calculate the change in absorbtion spectrum over time.
Modelling Software
The modelling was primarily conducted using two platforms; [http://www.mathworks.co.uk/products/matlab/ MATLAB®] and KaSiM. Together they provide the necessary tools to create an accurate model of our system.
They have been used succesfully by previous iGEM teams, notably the Edinburgh 2010 and 2011 teams, both of which one Best Model at the European Jamboree. This was the primary reason we chose the Kappa language to write our model in.
KaSiM is a [http://en.wikipedia.org/wiki/Stochastic stochastic] simulator that executes files written in [http://www.kappalanguage.org/ Kappa]. Kappa is a rule based modelling language for protein interaction networks. To download KaSiM or the manual please visit the KaSiM page.
KaSiM
KaSiMis a [http://en.wikipedia.org/wiki/Stochastic stochastic]simulator of rule based models written in the Kappa language. It is open source program that uses a variation of the Gillespie's algorithm to generate statistically correct trajectories. We advise you take the time to read the introduction to KaSiM in the KaSiM manual .
Kappa
Kappa is a high level rule-based language designed to describe protein interaction networks. Rule-based languages describe a system by a governing set of rules. There is no change in the configuration of the system without the execution of a rule. There are five fundamental parts to the kappa language; agents, tokens, rules, rates and observables. A quick introduction to these concepts is given below. For more detailed information we suggest visiting the [http://www.kappalanguage.org/ Kappa] website or for full examples [http://rulebase.org/ RuleBase] which offers a repository of complete rule based models. We can also recommend the 2012 Edinburgh iGEM team's Introduction to Kappa which proved useful to us early on.
Agents and tokens
Agents and tokens represent the constituent molecules of a system. Agents are quantified in terms of their population number and are characterised having binding sites. Whereas tokens are quantified in terms of their concentration and do not have binding sites.
Rules
Rules represent specific bio-chemical processes, and involve either tokens, agents or both. Each rule has an initial condition which must be satisfied for the rule to be valid. If the conditions are met, the rule is applied according to the rate.
Rates
Each rule has a specific rate constant which determines the number of times the rule is applied for each instance of the rule.
Observables
Observables determine the output data in terms of either agent count or token concentration.
MATLAB®
[http://www.mathworks.co.uk/products/matlab/ MATLAB®] is a high-level language and interactive environment for numerical computation, visualization, and programming. It can be used to analyse data, develop algorithms, and create models and applications. For guidance on using MATLAB® please see the [http://www.mathworks.co.uk/help/matlab/ mathworks documentation] .
The model
The model describes the absorption spectrum of the cell as a function of time and a given time invariant incident light spectrum. This is achieved by calculating the effect of the known light input on the individual light sensitive proteins. This information is then fed into a stochastic simulation of the biological pathways that computes how the population of sensors and pigments vary over time. The total absorption spectrum over time is given by the sum of the product of every protein's population and the corresponding absorption spectrum of the protein. In this section each of these steps in considered in the following subsections.
It will be helpful to read the theory page and the modelling software section of this page before reading further as an understanding of the biological pathways and software is important.
Sensor activation rate calculation
In the rule based model each sensor is thought of as a two-state binary system (on/off). In the on state it catalyses the phosphorylation of the primary intermediate, sending a signal to the downstream pathway. In the off state it does not.
Activation by light either turns a sensor on or off depending on the sensor species; red and blue light sensors are switched off when activated, while the green light sensor is switched on when activated. The activation rate is the forward rate of the rule that governs the state of a sensor. Its counterpart, the deactivation rate is the backward rate of the same rule and is constant. The activation rate is calculated as the integral over all space of the product of the sensor's frequency response with the frequency spectrum of the incident light.
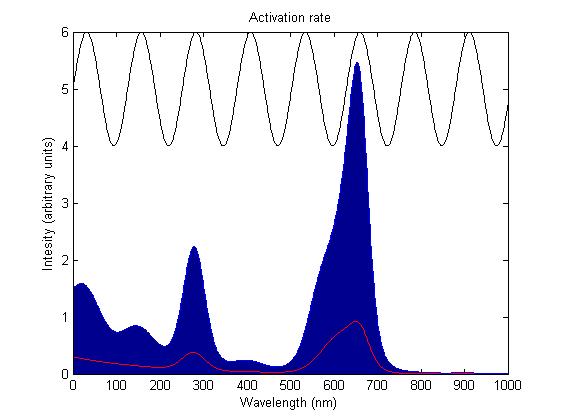
This has two important implications. Firstly it means that the sensor will not be activated by light that has no overlap between its spectrum and the frequency response of the sensor. Secondly there will be a saturation point at high intensity light where increasing the intensity will not appreciably affect the system as the activation rate will dominate the deactivation rate. These characteristics are representative of those of the light sensitive proteins being used.
Biological pathway simulation
Our stochastic model is comprised of red, green and a blue light activated pathways. Using multiple runs of the simulation, the stability of the tri-chromatic control system, its pigment balance, and image development time can all be tested in-silico. These have implications for future projects that might use a similar system and the model provides a platform for others to build on in the future.
Each pathway is independent in their signalling but shares the common resources of the cell. The independent variable is the activation rate of each light sensor species, this is determined by the incident light and effects the expression of pigments, the dependant variable.
The Kappa model describes in detail the pathway specific biochemistry and includes simplified descriptions of pathway related biochemistry. For example, the controlled transcription and translation of pigment proteins is described in full, but the constitutive transcription and translation of light sensor proteins is simplified. The rates were created to recreate experimental results recorded in literature. With experimental results this model can be readily updated to accurately describe our system.
All molecule species, with the exception DNA, are constitutively expressed. Molecule species apart from DNA, RNA polymerase and ribosomes have spontaneous decay rates, as variance during the time period of the system run has been taken as negligible. Phosphorylated species spontaneously dephosphorylation rate. Molecule species are represented as agents with the exceptions of the pigment proteins and ATP, which are modelled as concentrations of tokens.
The initial condition only contains DNA, RNA polymerase and ribosomes, and OmpR (which is naturally present in the E.coli), all other agents and tokens are produced by processes of transcription and translation. Constitutive expression of light sensors and intermediates are required before the cell can begin to respond to light.
Cell absorption spectrum calculation
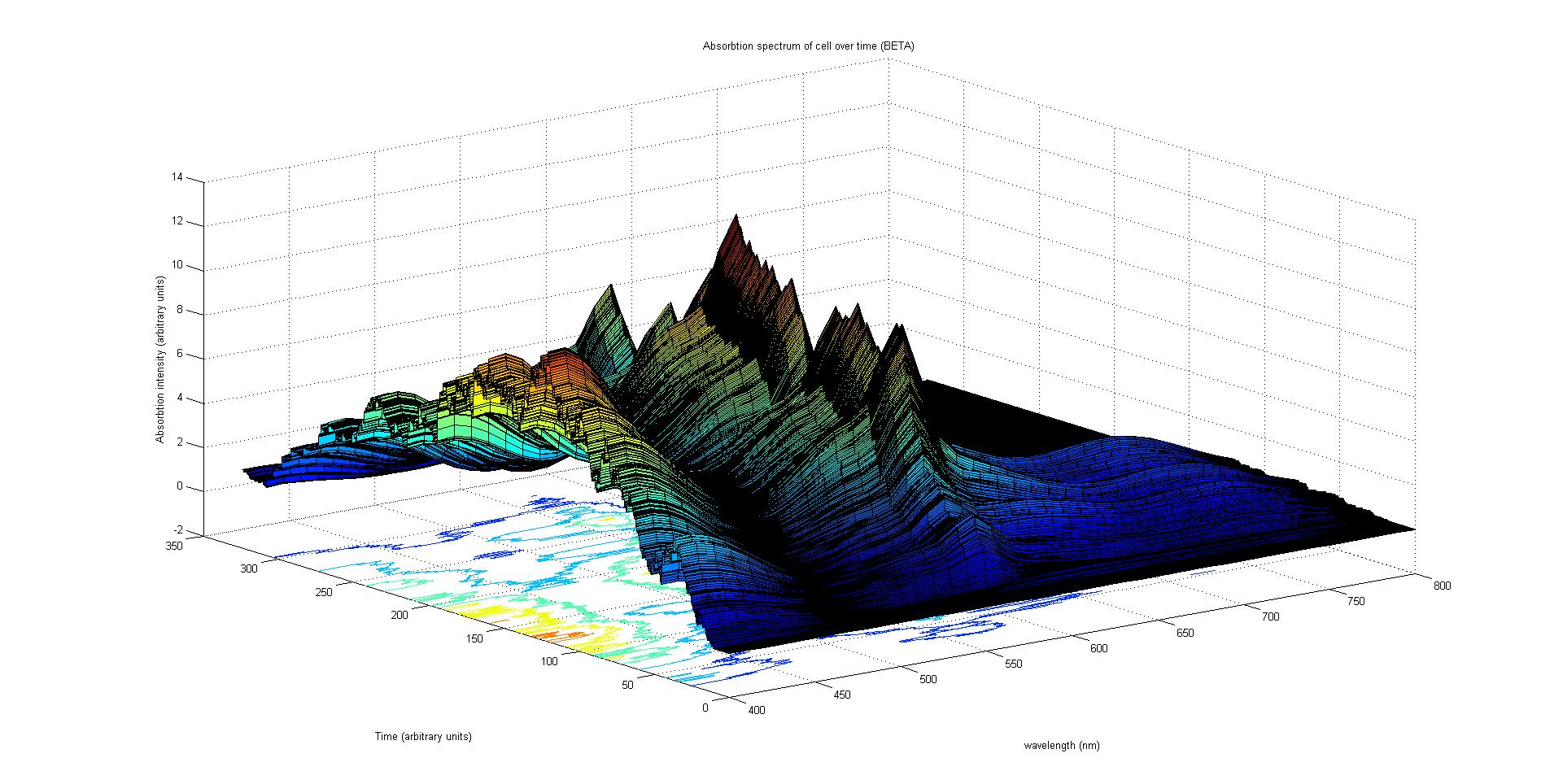
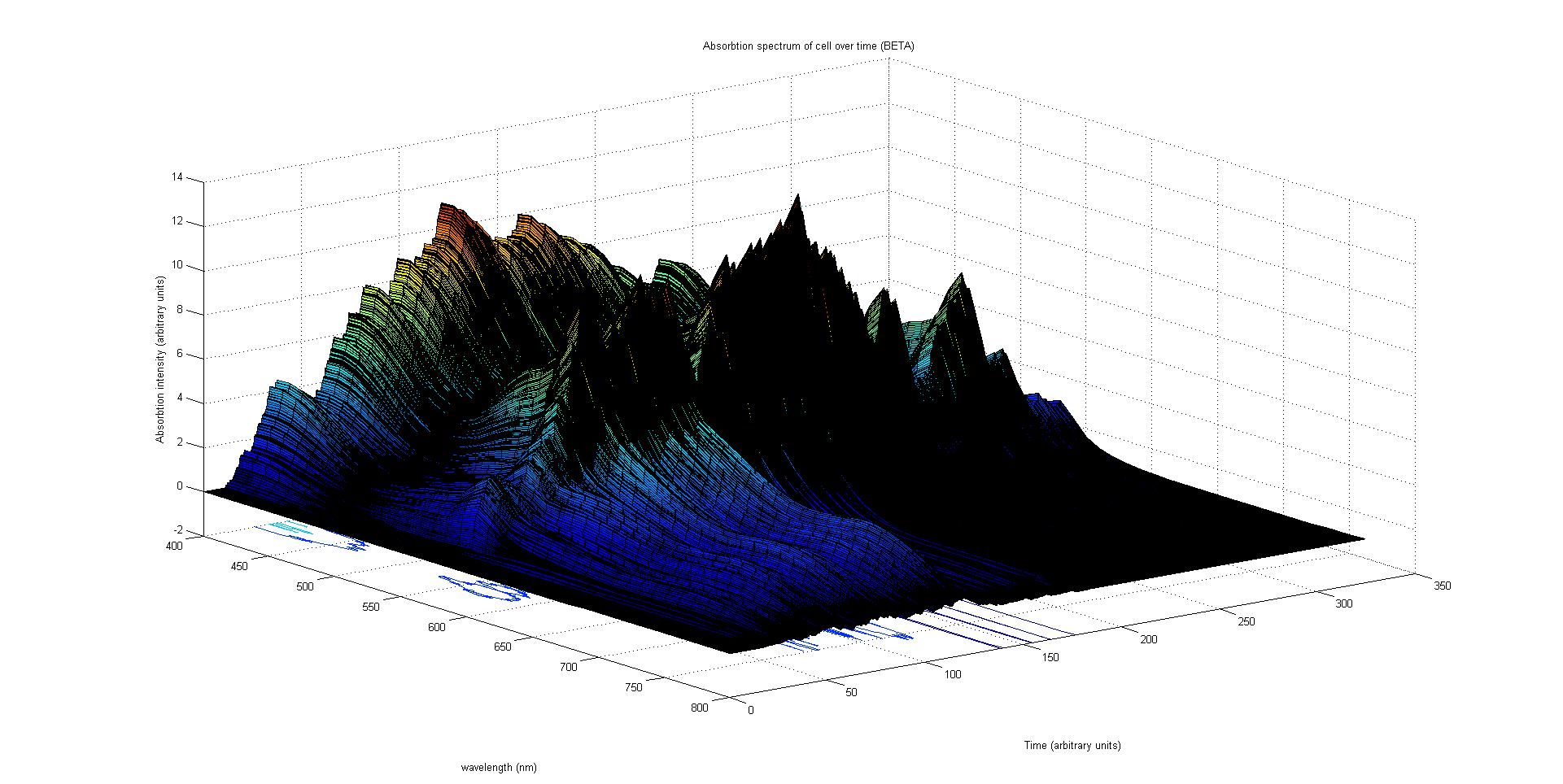
The final output of our model is a surface plot of absorption intensity as a function of time and wavelength. It reveals how the absorption spectrum and thus the colour of the cell changes with time. It also reveals how long it takes to reach equilibrium, the moment of greatest contrast and many other important features.
Each protein species has an associated absorption spectrum.
This multiplied by the population of the protein at a point in time gives the contribution of that protein to the total absorption spectrum of the cell at that moment. The sum of the contributions from each protein plus the absorption spectrum of the background cell gives the total absorption spectrum of the cell at that moment. This calculation repeated for every time step gives the total absorption of the cell over time.
Data sources
The model is based on data from literature and theoretical conjecture:
- The stochastic rates are ballpark figures for E.coli cells and are non specific.
- All absorbtion spectra do not have relative intensity all peak around 1 arbitrary unit.
- The light sensor absorption spectra are estimates of data taken from literature.
- The pigment sensor absorption spectra are estimates of the pigment absoption of colour printer ink.
- The operation of the light sensor is an idealisation of the true relationship between sensor activation and incident light
These will all contribute to the errors in our model but have been chosen to demonstrate our model in action. These can easily be replace with experimental results.
Results and analysis
All results in this section are obtained from the beta version of our model. In this section we will discuss the results that our model has produced and also how sound the model's operation is.
Results of model
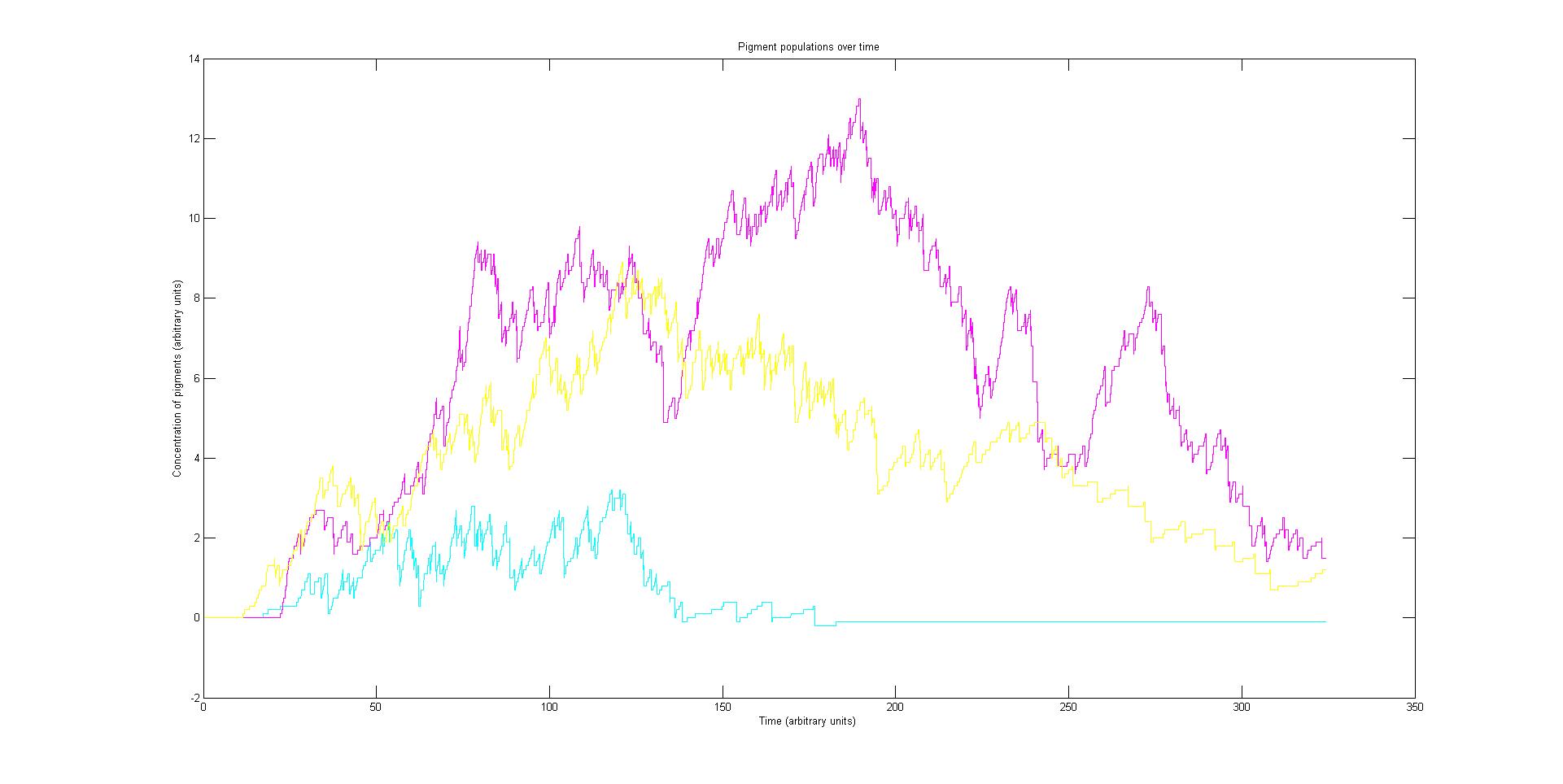
Preliminary results suggest that the mix of colours in any individual E.coli system is very unpredictable. The model was run multiple times with all sensors at the same activation rate. The mix of colours between separate simulations is very unstable. However the average for a cohort of cell systems is more stable. It is due to the relatively small number of DNA agents with which the RNAP interacts. This amplifies the effects of chance and causes the instability observed in the model.

This will serve reduce the spacial control of tri-chromatic control system in E.coli as a predictable average expression of a gene will require multiple individuals. For a biocamera this will reduce the pixel density and therefore the resolution. One solution to this potential problem could be to introduce many copies of the light regulated genes. This could also serve to prevent one pathway dominating by having greater numbers of less prolific light regulated genes to compensate.
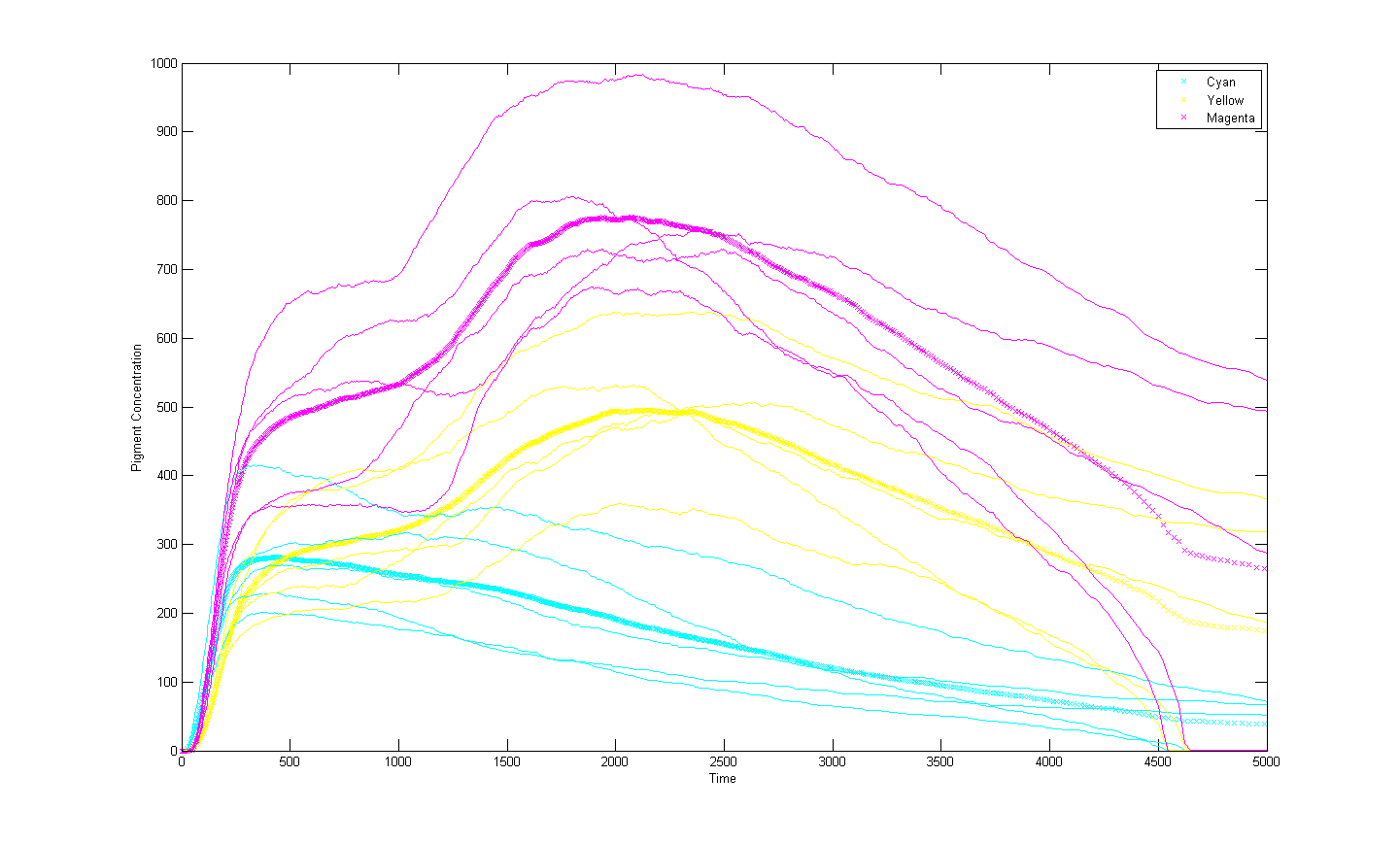
An increase in light regulated DNA has been shown to stabilise the system. However it is impractical and expensive to insert large amounts of DNA into individual bacteria with current methods.
User experience
The activation rate script is simple and deterministic. The results for activation rates are usually very high but that can be counterbalanced by either reducing the intensity of either the sensor absorption spectrum and/or the incident light. The results of this method have two important parrallels with the physical reality making it a working simplified model of sensor activation.
The Stochastic model is very sensitive to changes in rates and initial conditions. With very different behaviour resulting from only small pertubations. This is due to the stochastic nature of the model compounded by the small number of DNA agents as discussed before.
The absorption spectrum over time surface plot has to potential to produce accurate results. We have writen a script in MATLAB® that calculates the reflected light over time given the absorption over time and the incident light. It is possible to write a program that converts the reflected light spectrum over time into a colour over time. Regrettfuly we did not have the time to attempt such an edevour.
Sources of error
The model we have created is limited by the lack of experimental data to test and update it. As a result the data used in our model has a variety of sources and these indroduce many sources of error.
Firstly the rules describing the biochemistry of our cell addressed only the reactions unique to our pathway in detail and related reactions in a simplified manner. A small number of external conditions were addressed such as the competition for RNA polymerase and ribosomes between our pathway and other activity in the cell. This however means that the model does not take into account any interference from other cell activity. This is a source of error in our pathway simulation, however it is unlikely to the primary source of error.
Secondly the absorbtion spectra for all proteins and the background cell do not have their relative absorbtion intensities. Furthermore the spectra of the light sensors are approximations to data in literature, but the blue light sensor's absorbtion is that of LOVtap not YF1. LOVtap and YF1 are both light oxygen voltage blue light receptors and therefore we expect them to have a similar absorbtion spectrum. The spectra of the pigments are approximations of the standard cyan, magenta and yellow pigments used in colour printers. Therefore the pigment absorbtion spectra only represent the ideal colour of our pigments. This final point does not affect our rule-based stochastic simulation. The inaccuracy in our spectra causes an uncertainty in the calculation of activation rate and a greater uncertainty in the calculation of the cell's absorbtion.
Thirdly the nature of the activation of the light sensors is unknown and the method we use to calculate the activation rate is likely to be errenous. This introduces another source of error in the calculation of sensor activation rates.
Fourthly the rates in our rule-based model are ballpark figures taken from literature and are not specific to the reactions in our model. This is the primary source of error in our pathway simulation.
Conclusions
The multitude and magnitude of error sources in our model mean that no accurate conclusions about our synthetic biological system can be drawn from it.
However our preliminary results suggest that tri-chromatic control of gene expression in E.coli is very unstable for individual cells. This may be due to the small number of DNA blocks in our system amplifying the effect of chance. For large groups of E.coli the average expression is more predictable. This is to be expected in a stochastic system but the degree to which expression varies between individuals under identical circumstances surprised us. This suggests that instability will reduce the spatial control that our tri-chromatic system can have. It is possible that increasing the number of light regulated genes may reduce the instability and increase the potential spatial resolution.
The model itself is a good foundation for future multi-chromatic control modelling efforts. The data that feeds the model can easily be replace with experimental data. The kappa file is well layed out and comprehensible with some understanding of kappa syntax. We think that it has the potential to provide a useful tool for modelling similar systems or as a resource for others to build rule-based models of their own.
Future
Our model serves as a framework for modelling tri-chromatic control in E.coli. The foremost action to be taken in the future is the successful synthesis and testing of a working tri-chromatic control system. The experimental results could be used to update and test the model.
Improvements to the workings of the model include allowing the input signal to change with time. This means updating the rates in the kappa model while the model is being run by KaSiM. This requires either greater integration between KaSiM and mathematics programs like MATLAB or a more powerful mathematics suite in KaSiM. It is possible also to write a program the converts the cell's absorbtion spectrum over time into its colour over time. This is something that we did not have time to do.
Increasing the number of light regulated DNA in a cell reduces the instability. Perhaps in the future it will become affordable to genome integrate large numbers of light regulated genes to provide reliable and extremely high spatial control of bacteria using mulitple wavelengths.
Bibliography
Edinburgh iGEM 2010 https://2010.igem.org/Team:Edinburgh/
KaSim3 reference manual (release 3.4) Jérôme Feret and Jean Krivine1 KappaLanguage.org
J Mol Biol. 2011 Jan 14;405(2):315-24. doi: 10.1016/j.jmb.2010.10.038. Epub 2010 Oct 28. Multichromatic control of gene expression in Escherichia coli. Tabor JJ, Levskaya A, Voigt CA.
Department of Pharmaceutical Chemistry, University of California, San Francisco, CA 94158, USA.
J Mol Biol. 2012 Mar 2;416(4):534-42. doi: 10.1016/j.jmb.2012.01.001. Epub 2012 Jan 8. From dusk till dawn: one-plasmid systems for light-regulated gene expression. Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Möglich A. Source
Humboldt-Universität zu Berlin, Institut für Biologie, Biophysikalische Chemie, Invalidenstraße 42, 10115 Berlin, Germany.
277, 27 October 2011, Pages 77–87
Modular Modelling in Synthetic Biology: Light-Based Communication in E. coli ☆ The Second International Workshop on Interactions between Computer Science and BiologyDonal Stewart E-mail the corresponding author DemonSoft Ltd, Edinburgh, United Kingdom John Roger Wilson-Kanamori E-mail the corresponding author
School of Informatics, University of Edinburgh, Edinburgh, United Kingdom
Applications of the Kubelka-Munk Color Model to Xerographic Images Final Report Kristen Hoffman Center for Imaging Science Rochester Institute of Technology May 1998
Raw code
Activation rate calculation
%% Import data %%
%from Kappa fid = fopen('combined-out.out','r'); populations = importdata('combined-out.out'); fclose(fid);
%% Set the absorbtion spectra %%
%Wavelength domain of absorbtions in nm as row array x = [300:800]; X = numel(x); Wavelength = x;
%Time is a column arrays Time = populations.data(:,1); %takes the time in arbitrary units from the KaSiM simulation results T = numel(Time); %number of time steps
%Pigment spectra are made up of exponential decay and gaussian functions to %roughly match the graphs for printer ink absorbtion taken from %http://www.cis.rit.edu/research/thesis/bs/1998/hoffman/Thesis.html
cyanspectrum = 0.5.*exp(-(x-200)/50)+0.6.*exp(-((x/3-300/3).^2)/400)+exp(-((x/3-620/3).^2)/400)+0.8.*exp(-((x/3-730/3).^2)/400);;%frequency spectrum of cyan pigment cyanspectrum1 = repmat(cyanspectrum,T,1);%spectrum expanded into a matrix with 100001 rows
magentaspectrum = exp(-(x-200)/50)+0.2.*exp(-((x/3-410/3).^2)/400)+0.7.*exp(-((x/3-540/3).^2)/400)+0.5.*exp(-((x/2-570/2).^2)/50); magentaspectrum1 = repmat(magentaspectrum,T,1);
yellowspectrum = 1.8*exp(-(x-300)/200).*exp(-((x/3-415/3).^2)/400); yellowspectrum1 = repmat(yellowspectrum,T,1);
%Sensor spectra are made up of exponential decay and gaussian functions to %roughly match the graphs on UCL PG team although I believe the original %images are atributed to Edinburgh 2010 iGEM team
redspectrum = 0.3*exp(-x/200)+0.3*exp(-((x/1.5-370/2).^2)/640)+0.7.*exp(-((x/2-620/2).^2)/1000)+0.4.*exp(-((x/2-660/2).^2)/200); redspectrum1 = repmat(redspectrum,T,1);
greenspectrum = 0.5*exp(-(x-300)/50)+0.6*exp(-((x/1.5-370/1.5).^2)/600)+exp(-((x/2-540/2).^2)/1000); greenspectrum1 = repmat(greenspectrum,T,1);
bluespectrum = 0.2*exp(-(x-300)/200)+0.4*exp(-((x/1-380).^2)/600)+0.9.*exp(-((x-440).^2)/500)+0.6.*exp(-((x-475).^2)/100); bluespectrum1 = repmat(bluespectrum,T,1);
%% Activation rate calculation
input = 5+sin(x); %This is the incident light wavelength spectrum user defined
redresponse = redspectrum.*input; % The curve redresponse is the product of the absorption spectrum of the red light sensor with its population at each time step greenresponse = greenspectrum.*input; blueresponse = bluespectrum.*input;
redactivation = trapz(x,redresponse) %The area under the redresponse curve is the activation rate of red greenactivation = trapz(x,greenresponse) blueactivation = trapz(x,blueresponse)
dlmwrite('ra',redactivation); dlmwrite('ga',greenactivation); dlmwrite('ba',blueactivation);
Absorbtion of cell over time
%% This is a script for matlab %% Exeter iGEM 2013 Angus Laurenson %% This script has three functions: %Calculates the rate of activation for each sensor %Takes the output file from KaSim and plots the absorbtion spectrum of the cell over time %Plots the reflected light over time using the absorbtion spectrum over %time
%% Import data %%
%from Kappa fid = fopen('combined-out.out','r'); populations = importdata('combined-out.out'); fclose(fid);
%% Set the absorbtion spectra %%
%Wavelength domain of absorbtions in nm as row array x = [300:10:800]; X = numel(x); Wavelength = x;
%Time is a column arrays Time = populations.data(:,1); T = numel(Time);
%Pigment spectra are made up of exponential decay and gaussian functions to %roughly match the graphs for printer ink absorbtion taken from %http://www.cis.rit.edu/research/thesis/bs/1998/hoffman/Thesis.html
cyanspectrum = 0.5.*exp(-(x-200)/50)+0.6.*exp(-((x/3-300/3).^2)/400)+exp(-((x/3-620/3).^2)/400)+0.8.*exp(-((x/3-730/3).^2)/400);;%frequency spectrum of cyan pigment cyanspectrum1 = repmat(cyanspectrum,T,1);%spectrum expanded into a matrix with 100001 rows
magentaspectrum = exp(-(x-200)/50)+0.2.*exp(-((x/3-410/3).^2)/400)+0.7.*exp(-((x/3-540/3).^2)/400)+0.5.*exp(-((x/2-570/2).^2)/50); magentaspectrum1 = repmat(magentaspectrum,T,1);
yellowspectrum = 1.8*exp(-(x-300)/200).*exp(-((x/3-415/3).^2)/400); yellowspectrum1 = repmat(yellowspectrum,T,1);
%Sensor spectra are made up of exponential decay and gaussian functions to %roughly match the graphs on UCL PG team although I believe the original %images are atributed to Edinburgh 2010 iGEM team
redspectrum = 0.3*exp(-x/200)+0.3*exp(-((x/1.5-370/2).^2)/640)+0.7.*exp(-((x/2-620/2).^2)/1000)+0.4.*exp(-((x/2-660/2).^2)/200); redspectrum1 = repmat(redspectrum,T,1);
greenspectrum = 0.5*exp(-(x-300)/50)+0.6*exp(-((x/1.5-370/1.5).^2)/600)+exp(-((x/2-540/2).^2)/1000); greenspectrum1 = repmat(greenspectrum,T,1);
bluespectrum = 0.2*exp(-(x-300)/200)+0.4*exp(-((x/1-380).^2)/600)+0.9.*exp(-((x-440).^2)/500)+0.6.*exp(-((x-475).^2)/100); bluespectrum1 = repmat(bluespectrum,T,1);
%% Activation rate calculation
input = 5+sin(x);
redresponse = redspectrum.*input; greenresponse = greenspectrum.*input; bluesresponse = bluespectrum.*input;
% redactivation = trapz(x,redresponse); % greenactivation = trapz(x,greenresponse); % blueactivation = trapz(x,blueresponse);
% dlmwrite('ra',redactivation); % dlmwrite('ga',greenactivation); % dlmwrite('ba',blueactivation);
%% Calculation of absorbtion spectrum over time for each protein
cyanpop = populations.data(:,4);%populations are column arrays cyanpop = repmat(cyanpop,1,X);%populations are expanded into a matrix with 41 columns
magentapop = populations.data(:,3); magentapop = repmat(magentapop,1,X);
yellowpop = populations.data(:,2); yellowpop = repmat(yellowpop,1,X);
redpop = populations.data(:,5); redpop = repmat(redpop,1,X);
greenpop = populations.data(:,6); greenpop = repmat(greenpop,1,X);
bluepop = populations.data(:,7); bluepop = repmat(bluepop,1,X);
%% Multiplication of kappa population data with absorbtion spectra %%
cyan = cyanpop.*cyanspectrum1; magenta = magentapop.*magentaspectrum1; yellow = yellowpop.*yellowspectrum1;
red = redpop.*redspectrum1; green = greenpop.*greenspectrum1; blue = bluepop.*bluespectrum1;
%% Plot of the sum total absorbtion over time %%
absorbtion = cyan+magenta+yellow+red+green+blue;
pigments = cyan+magenta+yellow;
% plot(Time,cyanpop,'c',Time,magentapop,'m',Time,yellowpop,'y') % % title('Pigment populations over time') % xlabel('Time (arbitrary units)') % ylabel('Concentration of pigments (arbitrary units)')
% surfc(Wavelength,Time,pigments) % title('Absorbtion spectrum of cell over time (BETA)') % ylabel('Time (arbitrary units)') % xlabel('wavelength (nm)') % zlabel('Absorbtion intensity (arbitrary units)')
%% Plot of the reflected light given an incident light
%Original input incident % original = 20.*repmat(input,T,1); % % %White light incident % white = 20.*ones(T,X); % % incident = white; % reflected = incident-pigments; % % mesh(Wavelength,Time,reflected) % title('Reflected light spectrum of cell over time (BETA)') % ylabel('Time (arbitrary units)') % xlabel('wavelength (nm)') % zlabel('Light intensity (arbitrary units)')
%% Plots for wiki
plot(x,cyanspectrum,'c',x,magentaspectrum,'m',x,yellowspectrum,'y')
title('Pigment absorbtion spectra')
xlabel('Wavelength (nm)')
ylabel('Intensity (arbitrary units)')
clear
kappa
- Combined Model
- Henri Laakso;Angus Laurenson;Callum Vincent
- Observables ####
%obs: 'yellow' |Yellow| %obs: 'magenta' |magenta| %obs: 'cyan' |Cyan| %obs: 'red' Cph8() %obs: 'green' YF1() %obs: 'blue' CcaS()
- Signatures ####
- Agents
%agent: DNA(u,b,d,type~J23104~J23104L~K592006p1~K592006p2~B0034~B0034L~K592004~K592005~K592010~K592001~K592002~K592003p1~K592003p2~K592012~K608002~C0051~R0051p1~R0051p2~R0082p1~R0082p2~B0015~K592011~K592000~nonpath) %agent: RNA(d,b,u,type~B0034~K592004~K592005~K592010~B0015~K592001~K592002~K592003p1~K592003p2~K592012~K608002~C0051~R0051p1~R0051p2~R0051p3~R0051p4~K592000~K592011~nonpath) %agent: RNAP(dna,rna) %agent: RIB(rna,p,b) #Ribosome
%agent: YF1(b1,b2,type~on~off) %agent: FixJ(b,type~u~p) %agent: cl(b) #secondary intermediate %agent: CcaS(a,b,type~on~off) #green light detector %agent: CcaR(b~u~p) #primary intermediate %agent: OmpR(b,type~u~p) %agent: Cph8(b1,b2,type~on~off)
- Tokens
%token: ATP %token: ADP %token: Yellow %token: magenta %token: Cyan
- Variables ####
- Sensor activation rates
%var: 'receptor dimerization' 0.0036 %var: 'receptor dissociation' 360
%var: 'yf1 activation rate' 1 %var: 'ccas activation rate' 1 %var: 'cph8 activation rate' 1
- Phosphorylation and dephosphorylation rates
- %var: 'phosphorylation rate' 225
- %var: 'fixj phosphorylation rate' 'phosphorylation rate'
- %var: 'fixj phosphorylation base rate' 'phosphorylation rate'
- %var: 'CcaR phosphorylation rate' 'phosphorylation rate'
- %var: 'CcaR phosphorylation base rate' 'phosphorylation rate'
- %var: 'ompr phosphorylation rate' 'phosphorylation rate'
- %var: 'ompr phosphorylation base rate' 'phosphorylation rate'
- %var: 'atp phosphorylation rate' 'phosphorylation rate'
%var: 'fixj phosphorylation rate' 225 %var: 'fixj phosphorylation base rate' 2.5 %var: 'CcaR phosphorylation rate' 225 %var: 'CcaR phosphorylation base rate' 2.5 %var: 'ompr phosphorylation rate' 225 %var: 'ompr phosphorylation base rate' 2.5 %var: 'atp phosphorylation rate' 2.5
- %var: 'dephosphorylation' 2.25*(10^3)
- %var: 'CcaR dephosphorylation rate' 'dephosphorylation'
- %var: 'fixj dephosphorylation rate' 'dephosphorylation'
- %var: 'ompr dephosphorylation rate' 'dephosphorylation'
%var: 'CcaR dephosphorylation rate' 2.25*(10^2) %var: 'fixj dephosphorylation rate' 2.25*(10^2) %var: 'ompr dephosphorylation rate' 2.25*(10^2)
- Moving rates
%var: 'rnap moving rate' 10 %var: 'rib moving rate' 1.67
- Binding Rates
%var: 'general binding rate' 0.0022
%var: 'atp binding rate' 0.0022*|ATP| #0.5 %var: 'rnap binding rate' 0.0022 #0.0007 %var: 'rib binding rate' 0.0022 #0.000166 %var: 'fixj binding rate' 0.0022 #0.01 %var: 'ccar binding rate' 0.0022 #0.01 %var: 'cl binding rate' 0.0022 #0.01 #0.00996323269897635 edinburgh 2010's value %var: 'ompr binding rate' 0.0022 #0.01
- Falloff Rates
%var: 'general unbinding rate' 22.5
%var: 'rnap falloff rate' 'general unbinding rate'/20 %var: 'rib falloff rate' 'general unbinding rate'/20 #0.01 %var: 'fixj falloff rate' 'general unbinding rate'/20 #0.1 %var: 'ccar falloff rate' 'general unbinding rate'/20 #0.1 %var: 'CcaR u fall off rate' 'general unbinding rate'/20 #0.1 %var: 'ompr falloff rate' 'general unbinding rate'/20 #0.1
%var: 'cl fall off rate' 'general unbinding rate'/20 #0.001
- %var: 'cl fall off rate 1' #2.24 edinburgh 2010's value
- %var: 'cl fall off rate 2' #0.09 edinburgh 2010's value
- Biobrick Lengths
%var: 'B0034 length' 12 %var: 'B0015 length' 129 %var: 'C0051 length' 775 %var: 'K592000 length' 2238 %var: 'K592001 length' 2265 %var: 'K592002 length' 708 %var: 'K592003 length' 238 %var: 'K592004 length' 1131 %var: 'K592005 length' 621 %var: 'K592010 length' 699 %var: 'K592011 length' 702 %var: 'K592012 length' 681
- Transcription rates
%var: 'B0034 transcription rate' 200/'B0034 length' %var: 'B0015 transcription rate' 200/'B0015 length' %var: 'C0051 transcription rate' 200/'C0051 length' %var: 'K592000 transcription rate' 200/'K592000 length' %var: 'K592001 transcription rate' 200/'K592001 length' %var: 'K592002 transcription rate' 200/'K592002 length' %var: 'K592003 transcription rate' 200/'K592003 length' %var: 'K592004 transcription rate' 200/'K592004 length' %var: 'K592005 transcription rate' 200/'K592005 length' %var: 'K592010 transcription rate' 200/'K592010 length' %var: 'K592011 transcription rate' 200/'K592011 length' %var: 'K592012 transcription rate' 200/'K592012 length'
- Translation rates
%var: 'B0034 translation rate' 200/'B0034 length' %var: 'B0015 translation rate' 200/'B0015 length' %var: 'C0051 translation rate' 200/'C0051 length' %var: 'K592000 translation rate' 200/'K592000 length' %var: 'K592001 translation rate' 200/'K592001 length' %var: 'K592002 translation rate' 200/'K592002 length' %var: 'K592003 translation rate' 200/'K592003 length' %var: 'K592004 translation rate' 200/'K592004 length' %var: 'K592005 translation rate' 200/'K592005 length' %var: 'K592010 translation rate' 200/'K592010 length' %var: 'K592011 translation rate' 200/'K592011 length' %var: 'K592012 translation rate' 200/'K592012 length' %var: 'translation end rate' 200
- Degradation rates
%var: 'rnap degradation rate' 0 %var: 'rna degradation rate' 0.0058 %var: 'rib degradation rate' 0 %var: 'atp degradation rate' 0
%var: 'fixj degradation rate' 0.00115 %var: 'ccar degredation rate' 0.00115 %var: 'ompr degradation rate' 0.00115 %var: 'cl degredation rate' 0.00115
%var: 'yf1 degradation rate' 0.005 %var: 'ccas degradation rate' 0.005 %var: 'cph8 degradation rate' 0.005
%var: 'yellow degradation rate' 0.00001*|Yellow| %var: 'cyan degradation rate' 0.00001*|Cyan| %var: 'magenta degredation rate' 0.00001*|magenta|
- Initial Conditions ####
- Create DNA chains
%init: 1 DNA(u,b,d!1,type~J23104),DNA(u!1,b,d!2,type~B0034),DNA(u!2,b,d!3,type~K592004),DNA(u!3,b,d!4,type~B0015),DNA(u!4,b,d!5,type~K592005),DNA(u!5,b,d!6,type~B0034L),DNA(u!6,b,d!7,type~J23104L),DNA(u!7,b,d!8,type~K592006p1),DNA(u!8,b,d!9,type~K592006p2),DNA(u!9,b,d!10,type~B0034),DNA(u!10,b,d!11,type~K592010),DNA(u!11,b,d,type~B0015) %init: 1 DNA(u,b,d!1,type~J23104),DNA(u!1,b,d!2,type~B0034),DNA(u!2,b,d!3,type~K592001), DNA(u!3,d,b,type~B0015) #Constitutive gene for CcaS 2449 bp %init: 1 DNA(u,b,d!1,type~J23104),DNA(u!1,b,d!2,type~B0034),DNA(u!2,b,d!3,type~K592002), DNA(u!3,d,b,type~B0015) #Constitutive gene for CcaR 892bp %init: 1 DNA(u,d!1,b,type~K592003p1), DNA(u!1,b,d!2,type~K592003p2), DNA(u!2,b,d!3,type~B0034),DNA(u!3,b,d!4,type~C0051),DNA(u!4,b,d,type~B0015) #CcaR regulated gene for lambda cl 1154 bp %init: 1 DNA(u,d!1,b,type~R0051p1), DNA(u!1,b,d!2,type~R0051p2), DNA(u!2,b,d!3,type~B0034), DNA(u!3,b,d!4,type~K592012), DNA(u!4,b,d,type~B0015) #Lambda cl regulated gene for magenta pigment 871 bp %init: 1 DNA(u,b,d!1,type~R0082p1),DNA(u!1,b,d!2,type~R0082p2),DNA(u!2,b,d!3,type~B0034),DNA(u!3,b,d!4,type~K592011),DNA(u!4,b,d!5,type~B0015),DNA(u!5,b,d!7,type~J23104),DNA(u!7,b,d!8,type~B0034),DNA(u!8,b,d!9,type~K592000),DNA(u!9,b,d,type~B0015) %init: 100 DNA(u!0,b,d!1,type~nonpath),DNA(u!1,b,d!2,type~nonpath),DNA(u!2,b,d!3,type~nonpath),DNA(u!3,b,d!0,type~nonpath)
- Other initial conditions
%init: 700 RNAP() %init: 18000 RIB() #Ribosome %init: 50 OmpR(b,type~u)
- Tokens
%init: Cyan <- 0 %init: Yellow <- 0 %init: ATP <- 10000 %init: ADP <- 0 %init: magenta <- 0 #Magenta protein concentration
- Rules ####
- Degradation ##
'rnap degradation' RNAP() -> @ 'rnap degradation rate' 'RNA degradation' RNA() -> @ 'rna degradation rate' 'RIB degradation' RIB() -> @ 'rib degradation rate'
'FixJ degradation' FixJ() -> @ 'fixj degradation rate' 'CcaR degredation' CcaR() -> @ 'ccar degredation rate' 'OmpR degradation' OmpR() -> @ 'ompr degradation rate' 'cl degredation' cl() -> @ 'cl degredation rate'
'YF1 degradation' YF1() -> @ 'yf1 degradation rate' 'Ccas degradation' CcaS() -> @ 'ccas degradation rate' 'Cph8 degradation' Cph8() -> @ 'cph8 degradation rate'
'atp degradation' |1:ATP <-> |0:ATP @ 'atp degradation rate', 'atp phosphorylation rate' 'adp production' |0:ADP <-> |1:ATP @ 'atp degradation rate', 'atp phosphorylation rate'
'Yellow degradation' |0.5:Yellow -> |0:Yellow @ 'yellow degradation rate' 'Cyan degradation' |0.5:Cyan -> |0:Cyan @ 'cyan degradation rate' 'Magenta degradation' | 0.5:magenta -> | 0:magenta @ 'magenta degredation rate'
'FixJ dephosphorylation' FixJ(type~p) <-> FixJ(type~u) @ 'fixj dephosphorylation rate','fixj phosphorylation base rate' 'CcaR dephosphorylation' CcaR(b~p?) <-> CcaR(b~u?) @ 'CcaR dephosphorylation rate','CcaR phosphorylation base rate' 'OmpR dephosphorylation' OmpR(type~p) <-> OmpR(type~u) @ 'ompr dephosphorylation rate','ompr phosphorylation base rate'
- Sensors ##
- Sensor activation
YF1(type~off) -> YF1(type~on) @ 'yf1 activation rate' CcaS(type~off) -> CcaS(type~on) @ 'ccas activation rate' Cph8(type~off) -> Cph8(type~on) @ 'cph8 activation rate'
- Transcription factor phosphorylation at sensors
'OmpR phosphorylation at Cph8' Cph8(type~on),OmpR(b,type~u)|1:ATP -> Cph8(type~off),OmpR(b,type~p)|1:ADP @ 'ompr phosphorylation rate' 'FixJ phosphorylation at YF1' YF1(type~on),FixJ(b,type~u)|1:ATP -> YF1(type~off),FixJ(b,type~p)|1:ADP @ 'fixj phosphorylation rate' 'CcaR phosphorylation at CcaS' CcaS(a,b,type~on), CcaR(b~u) |1:ATP -> CcaS(a,b,type~on), CcaR(b~p) |1:ADP @ 'CcaR phosphorylation rate'
- Transcription ##
- Global rules
'DNA..RNAP' DNA(b!1),RNAP(dna!1,rna!2),RNA(d!2) -> DNA(b),RNAP(dna,rna),RNA(d) @ 'rnap falloff rate' 'DNA..FixJ' DNA(b!1),FixJ(b!1) -> DNA(b),FixJ(b) @ 'fixj falloff rate' 'DNA..CcaR' DNA(b!1),CcaR(b!1) -> DNA(b),FixJ(b) @ 'ccar falloff rate' 'DNA..OmpR' DNA(b!1),FixJ(b!1) -> DNA(b),FixJ(b) @ 'ompr falloff rate'
- Dephosphorylated CcaR detaches from the promoter that it is bound to
- 'CcaS u falls off' CcaR(b~u!1), DNA(u?,d?,b!1,type~K592003p1) -> CcaR(b~u), DNA(u?,d?,b,type~K592003p1) @ 'CcaR u fall off rate'
- Phosphrylated transcription factor binds to promoter region
'OmpR.R0082p1' DNA(b,type~R0082p1),OmpR(b,type~p) -> DNA(b!1,type~R0082p1),OmpR(b!1,type~p) @ 'ompr binding rate' 'FixJ.K592006p1' DNA(b,type~K592006p1),FixJ(b,type~p) -> DNA(b!1,type~K592006p1),FixJ(b!1,type~p) @ 'fixj binding rate' 'CcaR.K592003p1' DNA(b,type~K592003p1),CcaR(b~p) -> DNA(b!1,type~K592003p1), CcaR(b~p!1) @ 'ccar binding rate' 'cl.R0051p1' DNA(b,type~R0051p1),cl(b) -> DNA(b!1,type~R0051p1),cl(b!1) @ 'cl binding rate'
- rnap binds to promoter regions
'rnap.J23104' DNA(b,type~J23104),RNAP(dna) -> DNA(b!1, type~J23104),RNAP(dna!1) @ 'rnap binding rate' 'rnap.J23104L' DNA(b,type~J23104L),RNAP(dna) -> DNA(b!1,type~J23104L),RNAP(dna!1) @ 'rnap binding rate'
'rnap.R0082p2' DNA(b!3,d!2,type~R0082p1),DNA(u!2,b,type~R0082p2),OmpR(b!3),RNAP(dna) -> DNA(b!3,d!2,type~R0082p1),DNA(u!2,b!1,type~R0082p2),OmpR(b!3),RNAP(dna!1) @ 'rnap binding rate' 'rnap.K592006p2' DNA(b!3,d!2,type~K592006p1),DNA(u!2,b,type~K592006p2),FixJ(b!3),RNAP(dna) -> DNA(b!3,d!2,type~K592006p1),DNA(u!2,b!1,type~K592006p2),FixJ(b!3),RNAP(dna!1) @ 'rnap binding rate' 'rnap.K592003p2' DNA(d!1,b!3,type~K592003p1),DNA(u!1,b,type~K592003p2),CcaR(b!3),RNAP(dna) -> DNA(d!1,b!3,type~K592003p1),DNA(u!1,b!2,type~K592003p2),CcaR(b!3),RNAP(dna!2) @ 'rnap binding rate' 'rnap.R0051p2' DNA(b!3,d!1,type~R0051p1),DNA(u!1,b,type~R0051p2),cl(b!3),RNAP(dna) -> DNA(b!3,d!1,type~R0051p1),DNA(u!1,b!2,type~R0051p2),cl(b!3),RNAP(dna!2) @ 'rnap binding rate'
- Polymerase moves to first DNA block from promoter
'rnap moves from R0082p2' DNA(b!1,d!2,type~R0082p2),DNA(u!2,b),RNAP(dna!1) -> DNA(b,d!2,type~R0082p2),DNA(u!2,b!1),RNAP(dna!1) @ 'rnap moving rate' 'rnap moves from J23104' DNA(b!1,d!2,type~J23104),DNA(u!2,b),RNAP(dna!1) -> DNA(b,d!2,type~J23104),DNA(u!2,b!1),RNAP(dna!1) @ 'rnap moving rate' 'rnap moves from K592003p2' DNA(b!2,d!1,type~K592003p2),DNA(u!1,b),RNAP(dna!2) -> DNA(b,d!1,type~K592003p2),DNA(u!1,b!2),RNAP(dna!2) @ 'rnap moving rate' 'rnap moves from R0051p2' DNA(b!1,d!2,type~R0051p2),DNA(u!2,b),RNAP(dna!1) -> DNA(b,d!2,type~R0051p2),DNA(u!2,b!1),RNAP(dna!1) @ 'rnap moving rate' 'rnap moves from J23104L' DNA(b,d!2),DNA(u!2,b!1,type~J23104L),RNAP(dna!1) -> DNA(b!1,d!2),DNA(u!2,b,type~J23104L),RNAP(dna!1) @ 'rnap moving rate' 'rnap moves from K592006p2' DNA(b!1,d!2,type~K592006p2),DNA(u!2,b),RNAP(dna!1) -> DNA(b,d!2,type~K592006p2),DNA(u!2,b!1),RNAP(dna!1) @ 'rnap moving rate'
- Biobrick transcription
'B0034 transcription' DNA(b!1,d!2,type~B0034),DNA(u!2,b),RNAP(dna!1,rna) -> DNA(b,d!2,type~B0034),DNA(u!2,b!1),RNAP(dna!1,rna!3),RNA(d!3,b,u,type~B0034) @ 'B0034 transcription rate' 'B0015 transcription' DNA(b!1,type~B0015),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b,type~B0015),RNAP(dna,rna),RNA(d!3),RNA(d,b,u!3,type~B0015) @ 'B0015 transcription rate' 'C0051 transcription' DNA(b!1,d!2,type~C0051),DNA(u!2,b),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b,d!2,type~C0051),DNA(u!2,b!1),RNAP(dna!1,rna!4),RNA(d!3),RNA(d!4,u!3,type~C0051) @'C0051 transcription rate' 'K592000 transcription' DNA(b!1,d!2,type~K592000),DNA(u!2,b),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b,d!2,type~K592000),DNA(u!2,b!1),RNAP(dna!1,rna!4),RNA(d!3),RNA(d!4,b,u!3,type~K592000) @ 'K592000 transcription rate' 'K592001 transcription' DNA(b!1,d!2,type~K592001),DNA(u!2,b),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b,d!2,type~K592001),DNA(u!2,b!1),RNAP(dna!1,rna!4),RNA(d!3),RNA(d!4,b,u!3,type~K592001) @ 'K592001 transcription rate' 'K592002 transcription' DNA(b!1,d!2,type~K592002),DNA(u!2,b),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b,d!2,type~K592002),DNA(u!2,b!1),RNAP(dna!1,rna!4),RNA(d!3),RNA(d!4,b,u!3,type~K592002) @ 'K592002 transcription rate' 'K592004 transcription' DNA(b!1,d!2,type~K592004),DNA(u!2,b),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b,d!2,type~K592004),DNA(u!2,b!1),RNAP(dna!1,rna!4),RNA(d!3),RNA(d!4,b,u!3,type~K592004) @ 'K592004 transcription rate' 'K592005 transcription' DNA(b,d!2),DNA(u!2,b!1,type~K592005),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b!1,d!2),DNA(u!2,b,type~K592005),RNAP(dna!1,rna!4),RNA(d!3),RNA(d!4,b,u!3,type~K592005) @ 'K592005 transcription rate' 'K592010 transcription' DNA(b!1,d!2,type~K592010),DNA(u!2,b),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b,d!2,type~K592010),DNA(u!2,b!1),RNAP(dna!1,rna!4),RNA(d!3),RNA(d!4,b,u!3,type~K592010) @ 'K592010 transcription rate' 'K592011 transcription' DNA(b!1,d!2,type~K592011),DNA(u!2,b),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b,d!2,type~K592011),DNA(u!2,b!1),RNAP(dna!1,rna!4),RNA(d!3),RNA(d!4,b,u!3,type~K592011) @ 'K592011 transcription rate' 'K592012 transcription' DNA(b!1,d!2,type~K592012),DNA(u!2,b),RNAP(dna!1,rna!3),RNA(d!3) -> DNA(b,d!2,type~K592012),DNA(u!2,b!1),RNAP(dna!1,rna!4),RNA(d!3),RNA(d!4,b,u!3,type~K592012) @ 'K592012 transcription rate' 'B0034L transcription' DNA(b,d!2),DNA(u!2,b!1,type~B0034L),RNAP(dna!1,rna) -> DNA(b!1,d!2),DNA(u!2,b,type~B0034L),RNAP(dna!1,rna!3),RNA(d!3,b,u,type~B0034) @'B0034 transcription rate'
- Translation ##
- Global rules
'rib falloff' RNA(b!1),RIB(b!1) -> RNA(b),RIB(b) @ 'rib falloff rate'
- RIB binds to RIB binding site
'RIB.B0034' RNA(b,type~B0034),RIB(rna) -> RNA(b!1,type~B0034),RIB(rna!1) @ 'rib binding rate'
- Translation initiation
'rib moves from B0034' RNA(b!1,d!2,type~B0034),RNA(u!2,b),RIB(rna!1) -> RNA(d!2,b,type~B0034),RNA(u!2,b!1),RIB(rna!1) @ 'rib moving rate'
- Biobrick translation
'K592011 translation' RNA(b!1,d!2,type~K592011),RNA(u!2,b),RIB(rna!1) -> RNA(b,d!2,type~K592011),RNA(u!2,b!1),RIB(rna!1)|0.5:Cyan @ 'K592011 translation rate' 'K592000 translation' RNA(b!1,d!2,type~K592000),RNA(u!2,b),RIB(rna!1) -> RNA(b,d!2,type~K592000),RNA(u!2,b!1),RIB(rna!1),Cph8(b1,b2,type~off) @ 'K592000 translation rate' 'K592001 translation' RNA(b!1,d!2,type~K592001),RNA(u!2,b),RIB(rna!1) -> RNA(b,d!2,type~K592001),RNA(u!2,b!1),RIB(rna!1),CcaS(a,b,type~off) @ 'K592012 translation rate' 'K592002 translation' RNA(b!1,d!2,type~K592002),RNA(u!2,b),RIB(rna!1) -> RNA(b,d!2,type~K592002),RNA(u!2,b!1),RIB(rna!1),CcaR(b~u) @ 'K592002 translation rate' 'K592004 translation' RNA(b!1,d!2,type~K592004),RNA(u!2,b),RIB(rna!1) -> RNA(b,d!2,type~K592004),RNA(u!2,b!1),RIB(rna!1),YF1(b1,b2,type~off) @ 'K592004 translation rate' 'K592005 translation' RNA(b!1,d!2,type~K592005),RNA(u!2,b),RIB(rna!1) -> RNA(b,d!2,type~K592005),RNA(u!2,b!1),RIB(rna!1),FixJ(b,type~u) @ 'K592005 translation rate' 'K592010 translation' RNA(b!1,d!2,type~K592010),RNA(u!2,b),RIB(rna!1) -> RNA(b,d!2,type~K592010),RNA(u!2,b!1),RIB(rna!1)|0.5:Yellow @ 'K592010 translation rate' 'K592012 translation' RNA(b!1,d!2,type~K592012),RNA(u!2,b),RIB(rna!1) -> RNA(b,d!2,type~K592012),RNA(u!2,b!1),RIB(rna!1)|0.5:magenta @ 'K592012 translation rate' 'C0051 translation' RNA(b!1,d!2,type~C0051),RNA(u!2,b),RIB(rna!1) -> RNA(b,d!2,type~C0051),RNA(u!2,b!1),RIB(rna!1),cl(b) @ 'C0051 translation rate'
- Translation Completion
'RIB.B0015' RNA(b!1,type~B0015),RIB(rna!1) -> RNA(b,type~B0015),RIB(rna) @ 'translation end rate'
- Constitutive Expressions ####
- constitutive rates
%var: 'rnap move rate' 10 %var: 'rib move rate' 10 %var: 'rib fall rate' 1 %var: 'ompr nonpath binding rate' 0.001
'rnap.nonpath' DNA(b,type~nonpath),RNAP(dna) -> DNA(b!1,type~nonpath),RNAP(dna!1) @ 'rnap binding rate' 'ompr.nonpath' DNA(b,type~nonpath),OmpR(b,type~p) -> DNA(b!1,type~nonpath),OmpR(b!1,type~p) @'ompr nonpath binding rate'
'rnap free moves' DNA(b!1,d!2,type~nonpath),DNA(u!2,b,type~nonpath),RNAP(dna!1,rna) -> DNA(d!2,b,type~nonpath),DNA(u!2,b!1,type~nonpath),RNAP(dna!1,rna!3),RNA(d!3,b,u,type~nonpath) @'rnap move rate' 'rnap bound moves' DNA(b!1,d!2,type~nonpath),DNA(u!2,b,type~nonpath),RNAP(dna!1,rna!4),RNA(d!4,u?) -> DNA(b,d!2,type~nonpath),DNA(u!2,b!1,type~nonpath),RNAP(dna!1,rna!3),RNA(d!4,u?),RNA(d!3,b,u!4,type~nonpath) @'rib binding rate'
'rib.nonpath' RNA(b,type~nonpath),RIB(rna) -> RNA(b!1,type~nonpath),RIB(rna!1) @ 'rib binding rate' 'rib move' RNA(b!1,d!2,type~nonpath),RNA(u!2,b,type~nonpath),RIB(rna!1) -> RNA(b,d!2,type~nonpath),RNA(u!2,b!1,type~nonpath),RIB(rna!1) @'rib move rate' 'rib fall' RNA(b!1,d!2,type~nonpath),RNAP(rna!2),RIB(rna!1) -> RNA(b,d!2,type~nonpath),RNAP(rna!2),RIB(rna) @ 'rib fall rate'
 "
"