Team:Newcastle/Parts/HBsu-fp
From 2013.igem.org

Contents |
Hbsu-xFP Fusion
Purpose and Justification
Bacilluis subtilis contains HBsu, a non-specific DNA binding protein which is a homologue to eukaryotic histones. This protein is encoded by the gene hbs gene which is 263bp in length and involved not only in DNA binding but also SRP, DNA repair, HR, and pre-secretory protein translocation (Kouji, et al., 1999). It binds to DNA by forming homodimer.
We plan to conjugate the hbs gene with fluorescent protein (RFP/GFP) in order to fluorescently tag the bacterial chromosome. Each bacterium will only contain one or the other. We plan to regulate their expression through an IPTG induce promoter (Pspac).
This BioBrick the fusion of two L-form B. subtilis, and aims to improve existing phenotype via genome shuffling. This can be visualised through fluorescent microscopy, if a colour mosaic is formed we can confidently claim that recombination has occurred.
Assembly
HBsu-(x)FP
BioBrick encodes a fusion protein HBsu-(x)FP protein by 10 amino acids flexible linker (BBa_K105012).
The first version of the BioBrick have superfolder GFP (BBa_E0040)conjugated to the HBsu.
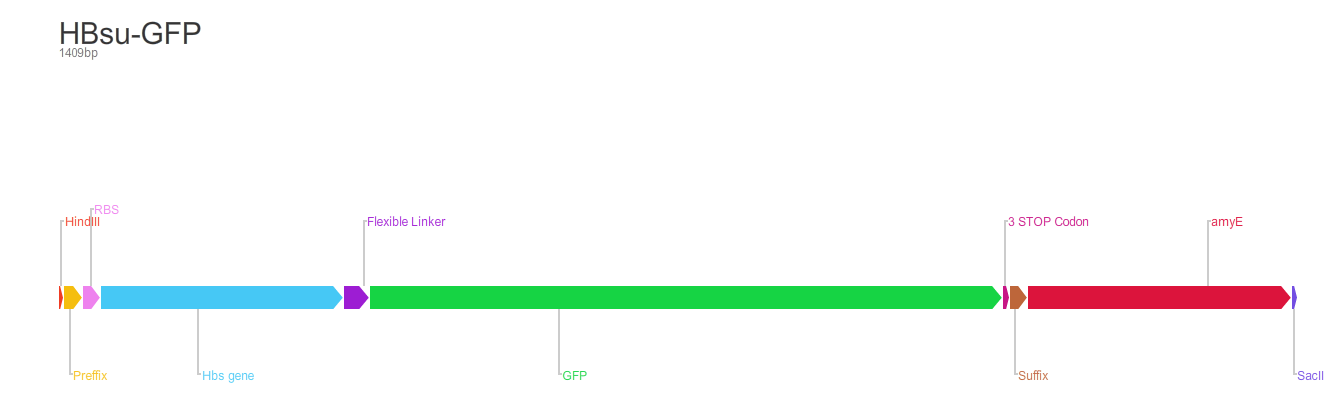
The second version of the BioBrick have RFP (BBa_E1010) conjugated to the HBsu.

Integration
Testing and Characterisation
 "
"