From 2013.igem.org
Aug 29
Colony PCR
No name
| Sample | base pair
|
| 8/27 Pcon-pT181 antisense-(1) | 542
|
| 8/26 pT181 attenuator-DT-(1) | 738
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | 8/27 Pcon-pT181antisense-1
|
| 3 | 8/26 pT181attenuator-DT-1
|
| 4 | 100bp ladder
|

Electrophoresis
No name
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | 8/21 RBS-lysis2-9(Colony PCR prodution)
|
| 3 | 8/21 RBS-lysis2-8(Colony PCR prodution)
|
| 4 | 8/21 RBS-lysis2-7(Colony PCR prodution)
|
| 5 | 8/21 RBS-lysis2-10(Colony PCR prodution)
|
| 6 | 8/21 RBS-lysis2-11(Colony PCR prodution)
|
| 7 | 8/21 RBS-lysis2-12(Colony PCR prodution)
|
| 8 | 8/21 RBS-lysis2-13(Colony PCR prodution)
|
| 9 | 8/21 RBS-lysis2-14(Colony PCR prodution)
|
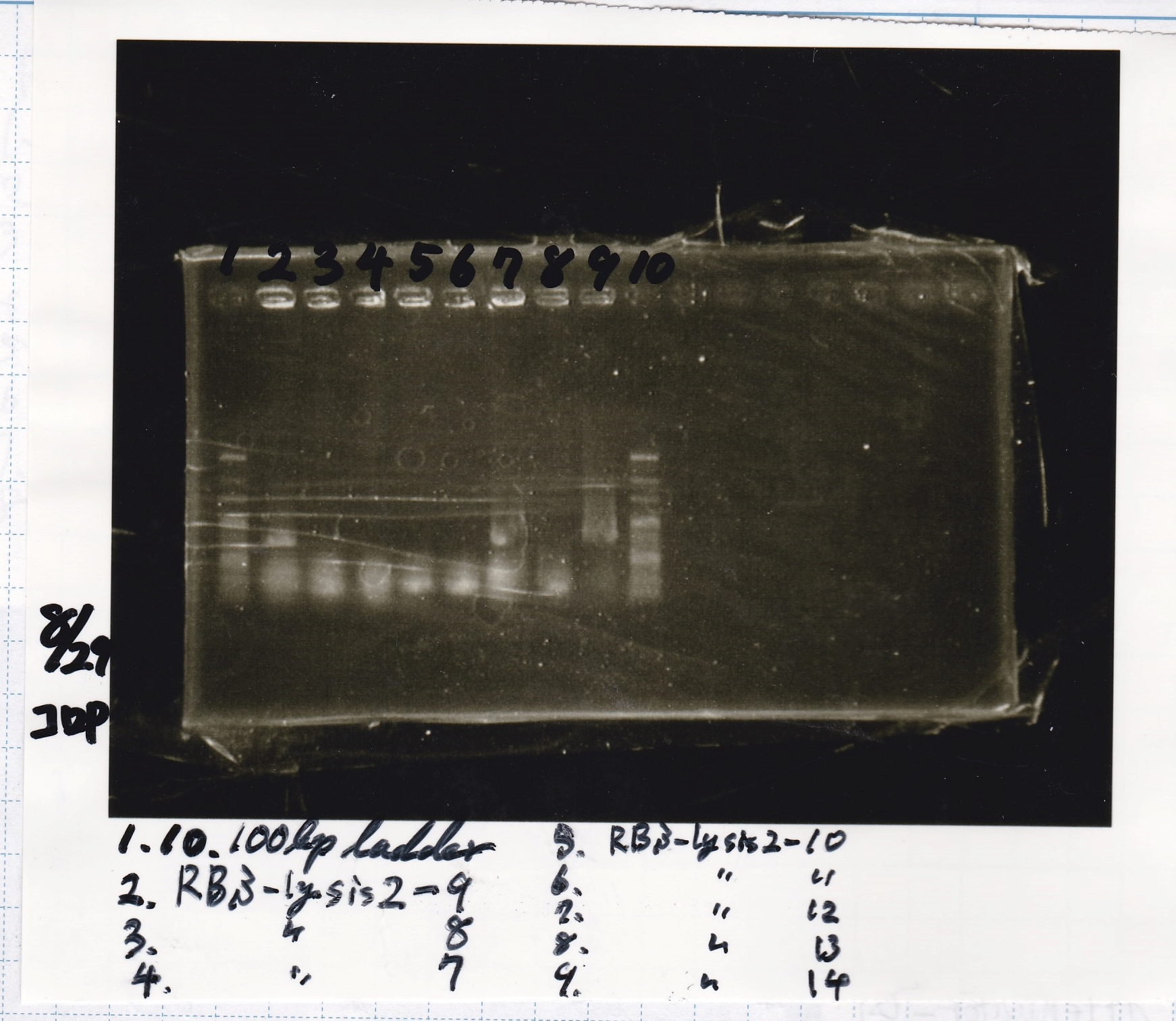
Miniprep
No Name
| DNA | concentration [µg/mL] | 260/280 | 260/230
|
| Plac | 153.7 | 2.03 | 2.10
|
| Pλ-luxⅠ-(1) | 76.5 | 2.07 | 2.06
|
| Pλ-luxⅠ-(2) | 86.4 | 2.08 | 2.10
|
| DT | 166.2 | 1.65 | 2.12
|
| J23100 | 190.1 | 1.68 | 2.34
|
Liquid Culture
No name
| Sample | medium
|
| 8/21 RBS-Lysis2-9 | Plusgrow medium(+Amp)
|
| 8/21 RBS-Lysis2-12 | Plusgrow medium(+Amp)
|
| 8/21 RBS-Lysis2-14 | Plusgrow medium(+Amp)
|
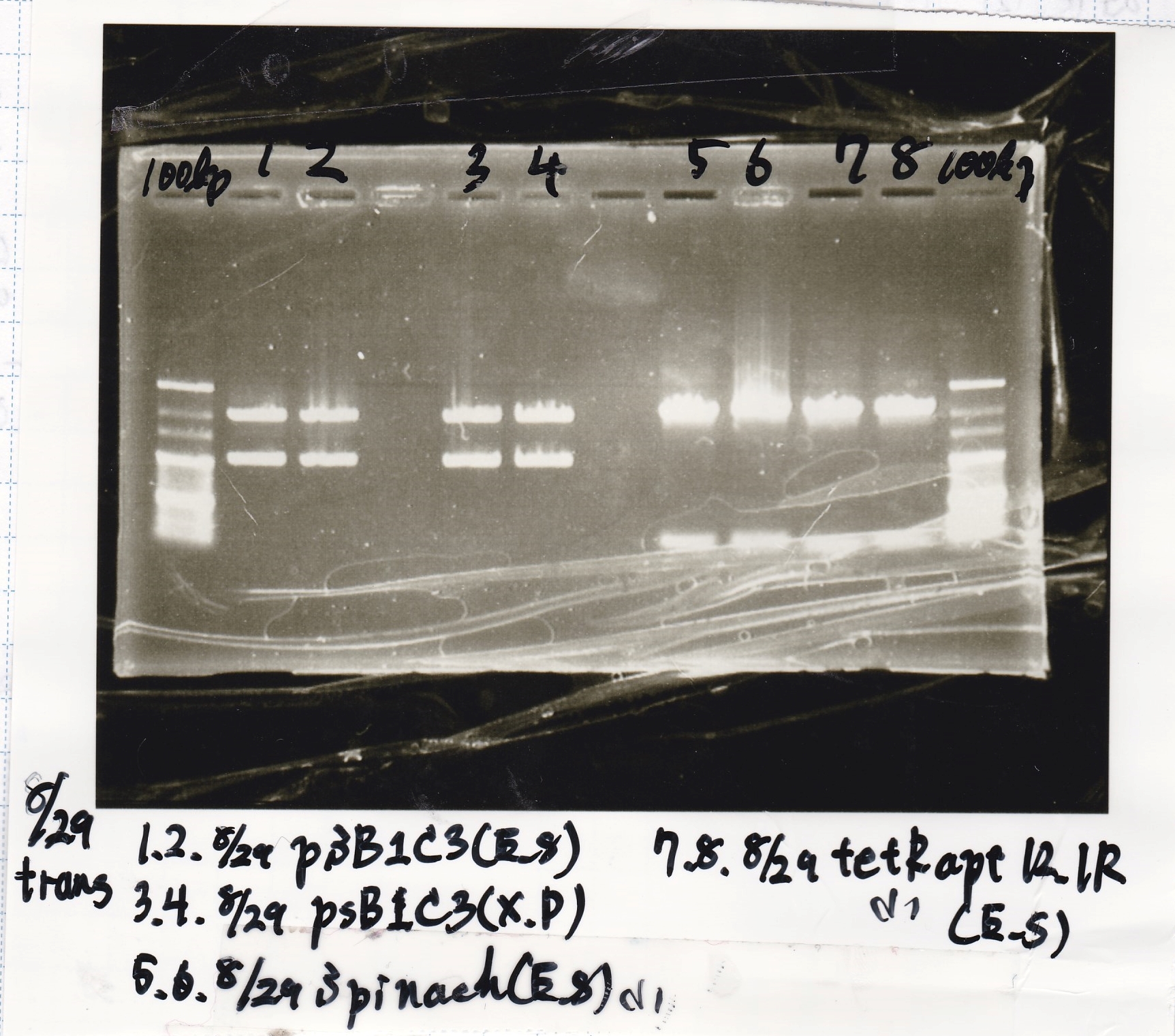
Restriction Enzyme Digestion
No name
| | 8/22 psB1C3-(2) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2 cuts | 8µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 17.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 7.9µL | 10µL
|
| 2 cuts | 8µL | 0µL | 0µL | 0.5µL | 0.5µL | 0µL | 3µL | 0.3µL | 17.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
| | 8/21 tRMA-spinach(1) | XbaI | SpeI | BufferB | BSA | MilliQ | total
|
| 2 cuts | 4µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 21.7µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
| | 8/21 tetR aptamer12-1R(1) | EcoRI | SpeI | BufferB | BSA | MilliQ | total
|
| 2 cuts | 3µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 22.7µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 0.1µL | 8.5µL | 10µL
|
| | 8/21 PT181 attenuator-(2) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2 cuts | 4µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 21.7µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.4µL | 10µL
|
| 2 cuts | 4µL | 0µL | 0µL | 0.5µL | 0.5µL | 0µL | 3µL | 0.3µL | 21.7µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
| | 8/21 PT181 antisense-(2) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2 cuts | 10µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 15.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 7.9µL | 10µL
|
| 2 cuts | 10µL | 0µL | 0µL | 0.5µL | 0.5µL | 0µL | 3µL | 0.3µL | 15.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
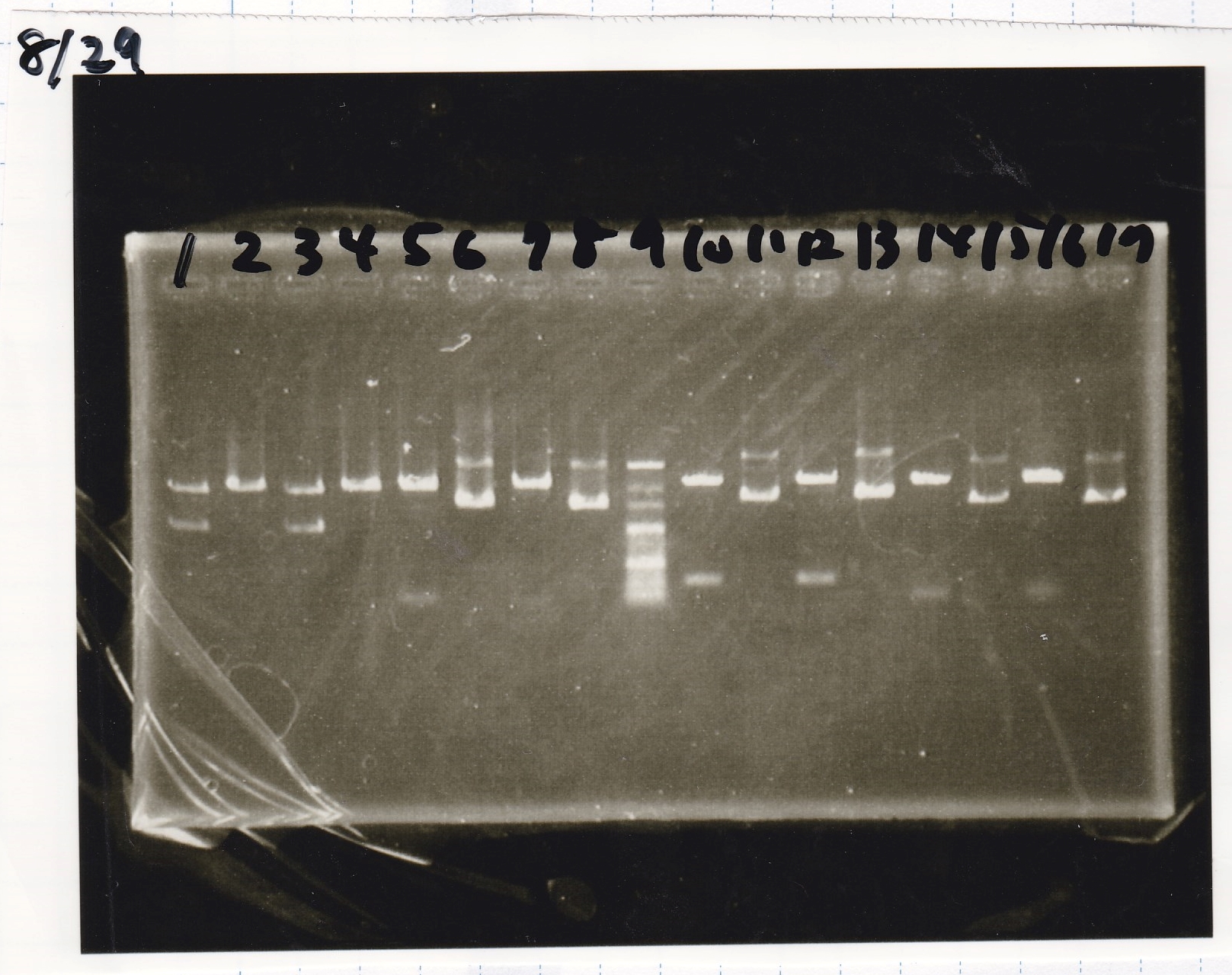
Transformation
No Name
| Name | Sample | Competent Cells | Total | Plate
|
| 8/27 pT181 attenuator-DT | 2µL | 20µL | 22µL | CP
|
| 8/27 RBS-lysis3-DT | 2µL | 20µL | 22µL | CP
|
| 8/28 RBS-lysis3-DT | 2µL | 20µL | 22µL | CP
|
| 8/28 Pcon-RBS-GFP-DT-Pcon-RBS-LuxR-DT | 2µL | 20µL | 22µL | Amp
|
| 8/28 RBS-lysis1-DT | 2µL | 20µL | 22µL | CP
|
| 8/28 Plux-RBS-GFP-DT | 2µL | 20µL | 22µL | CP
|
| 8/28 Spinach-DT | 2µL | 20µL | 22µL | CP
|

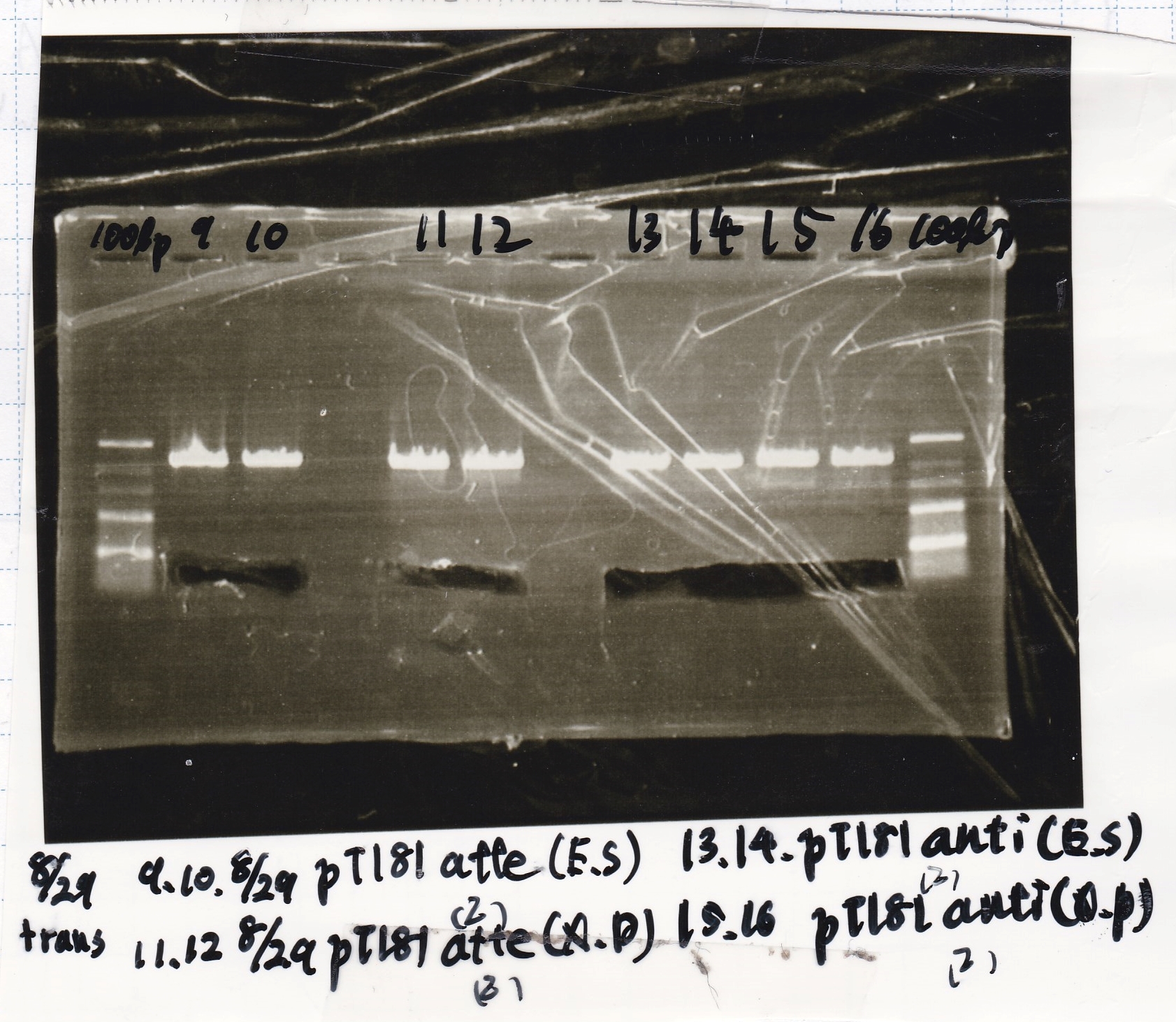
File:Igku Aug29 Transformation()after
No name
| Lane | DNA | Enzyme1 | Enzyme2
|
| 100bp ladder | -- | --
|
| 9 | 8/29 pT181attenuator(2) | EcoRI | SpeI
|
| 10 | 8/29 pT181attenuator(2) | EcoRI | SpeI
|
| 11 | 8/29 pT181attenuator(2) | XbaI | PstI
|
| 12 | 8/29 pT181attenuator(2) | XbaI | PstI
|
| 13 | pT181antisense(2) | EcoRI | SpeI
|
| 14 | pT181antisense(2) | EcoRI | SpeI
|
| 15 | pT181antisense(2) | XbaI | PstI
|
| 16 | pT181antisense(2) | XbaI | PstI
|
| 100bp ladder | -- | --
|



 "
"




