Team:ZJU-China/Project/GhostSensor/Results
From 2013.igem.org
Ghost Sensor: Result
Contents |
Characterize the function of protein E
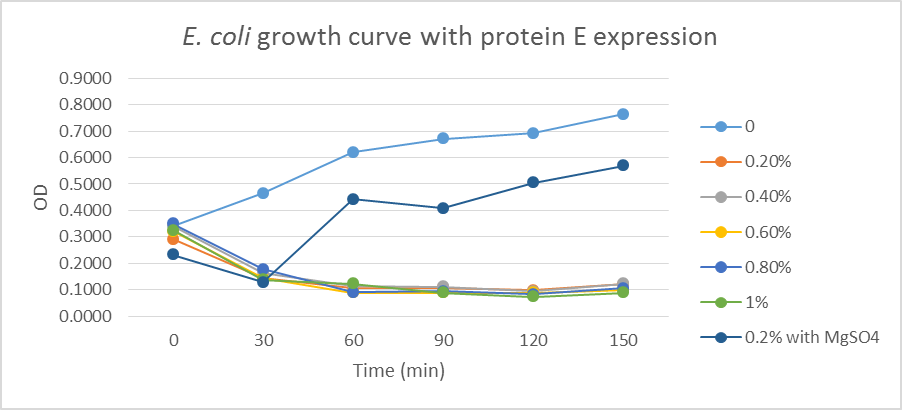
1.1 Protein E was put under the control of pBAD promoter. We recorded the growth curve of transformed E.coli cells treated with different concentrations of L-arabinose varying from 0.2% to 1%. MgSO4 has been reported to allow the expression of protein E but inhibit lysis. MgSO4 was added 30min before the induction according to previous works by others. The results showed that even as low as 0.2% L-arabinose can efficiently induce the lysis E.coli.
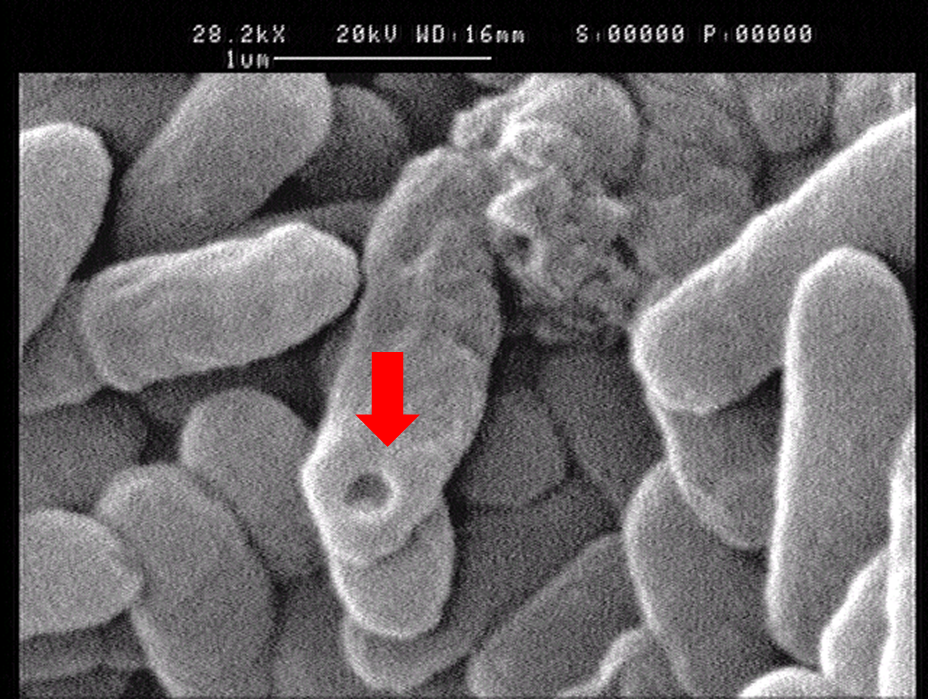
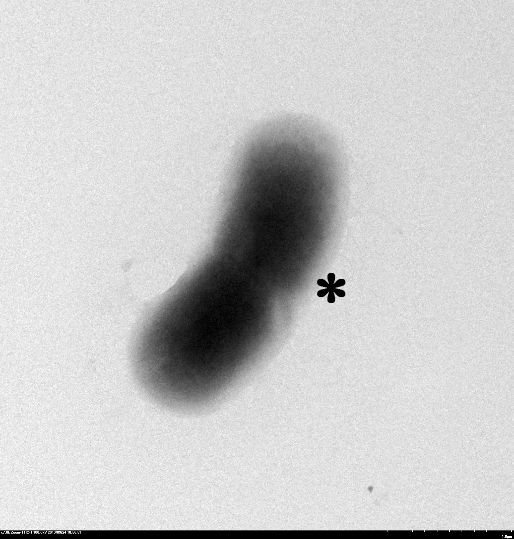
1.2 The lysed products were viewed under SEM and TEM to confirm the formation of tunnels. MgSO4 allows production of protein E, but inhibits lysis. 150min later cells were collected by centrifugation and resuspended in water causing immediate lysis by osmotic shock. This procedure would produce holes approximately equal in size to the diameter of the cell.
Left: SEM picture of protein E lysed E.coli.
Right: SEM picture of protein E lysed E.coli with MgSO4 protocol. The tunnel is relatively bigger than the left one.
TEM showing protein E lysed E.coli
Characterize the function of 29-mer and 15-mer thrombin aptamers
Detect thrombin using membrane scaffold FA and FB
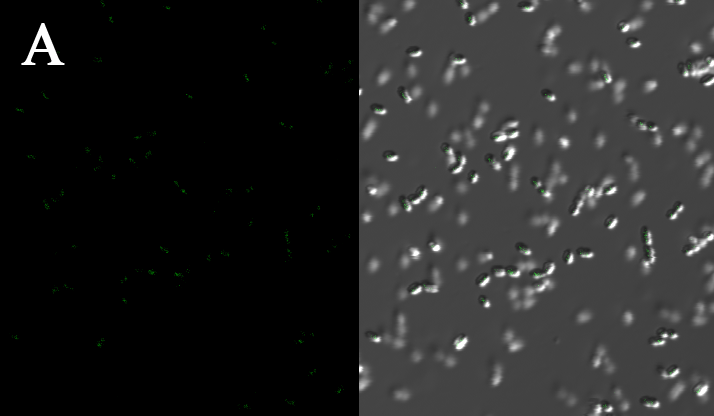
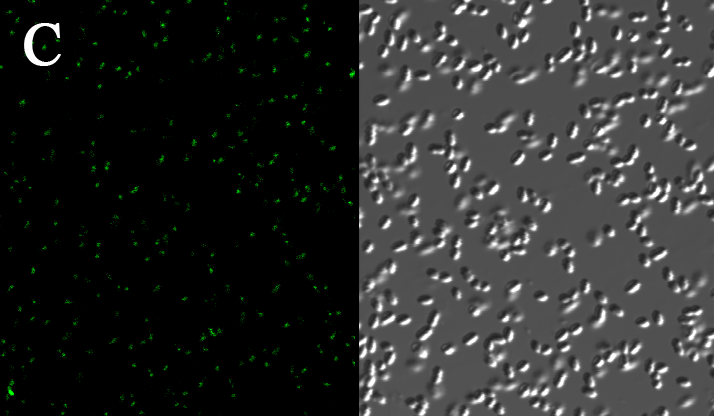
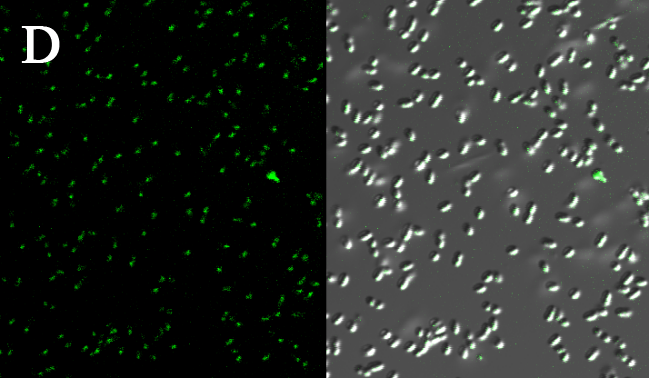
We expressed the first set of membrane scaffolds with split EGFP fragments FA and FB fused to the periplasmic end. Then we tested the function of our membrane scaffolds by adding two sets of aptamers with different linker length. The concentration of thrombin in the solution was adjusted to 0.2nM. 29-mer aptamer and 15-mer aptamer efficiently induced dimerization of membrane scaffolds (Figure C, D versus A) and led to green fluorescence emission under confocal microscopy.
Fluorescence with or without aptamers. BL21 were co-transformed with FA, FB and protein E. When the OD-600 value reached 0.4, 0.4% was added to induce the expression of protein E. 2h later cells were harvested and washed two times with PBS. 5µL of bacteria were resuspended in 200 µL PBS, and a final concentration of 0.2nM thrombin was added to the solution. Different aptamers were also included in this point. The reaction system was incubated with shaking in room temperature for 30 min and then green fluorescence was viewed under confocal microscopy. Only very weak green fluorescence was visible when no aptamers were added to the reaction system (A). Biotinylated thrombin can readily induce membrane scaffolds dimerization because there were typically 1-4 biontin in one molecule leading to scaffolds crosslinking (B). 15-mer and 29-mer aptamer with short (C) or long (D) linkers were both effective in inducing crosslinking. But it seemed that long linkers (D) is more potent than short linker (C) in this system.
 "
"