Reporter Team
Introduction
In order to facilitate the development of new antibiotics, we want to build an “Antibiotic Detector” that can identify substances influencing the homeostasis of cyclic-di-AMP (c-di-AMP). c-di-AMP is an essential signal nucleotide in most Gram-positive bacteria. It is not found in humans and several Gram-negative bacteria lack this compound, as well, including E. coli. For more information see Our Project overview.
For constructing a c-di-AMP detector, we need sensors for c-di-AMP and in order to visualize the output of the sensors, we need a reporter.
DarR reporter system ([http://parts.igem.org/Part:BBa_K1045017 part BBa_K1045017]):
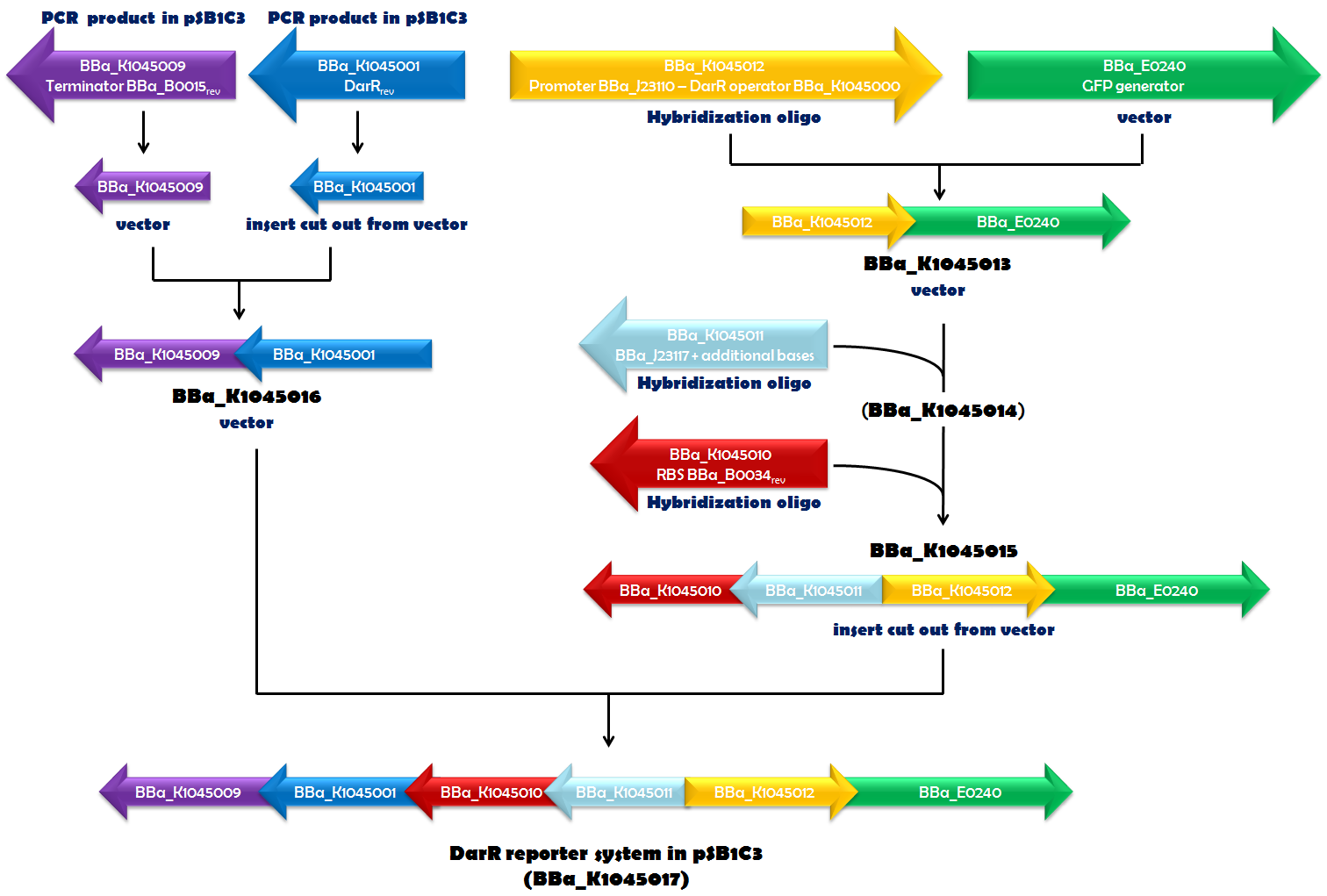
Recently, in Mycobacterium smegmatis, a transcriptional repressor (DarR) was identified. It can bind to a specific DNA sequence, called the DarR operator. The binding efficiency of DarR is strongly enhanced by c-di-AMP. Since DarR is a c-di-AMP sensor, we intended to use it for our reporter system. We cloned DarR into pSB1C3 ([http://parts.igem.org/Part:BBa_K1045001 BBa_K1045001]) and also constructed the operator as a Biobrick ([http://parts.igem.org/Part:BBa_K1045000 BBa_K1045000]). We placed the DarR operator between a strong promoter ([http://parts.igem.org/Part:BBa_J23110 BBa_J23110]) and the GFP generator [http://parts.igem.org/Part:BBa_E0240 BBa_E0240]. In the very same plasmid but oriented in the opposite direction, we assembled a DarR expression unit. DarR expression was driven by a weak promoter based on [http://parts.igem.org/Part:BBa_J23117 BBa_J23117] and terminated by [http://parts.igem.org/Part:BBa_K1045009 BBa_K1045009]. This terminator is based on [http://parts.igem.org/Part:BBa_B0015 BBa_B0015], which is part of [http://parts.igem.org/Part:BBa_E0240 BBa_E0240], as well, and should ensure that the transcription of DarR and GFP did not influence each other. The RBS [http://parts.igem.org/Part:BBa_K1045010 BBa_K1045010] (derived from [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]) was used as an RBS for DarR mRNA translation. Assembly of all those parts (Fig. 1.1) required a complex cloning process which finally resulted in part [http://parts.igem.org/Part:BBa_K1045017 BBa_K1045017], the DarR reporter system (Fig. 1.2). E. coli was then transformed with this construct to obtain the in vivo screening system for antibiotics directed against c-di-AMP.
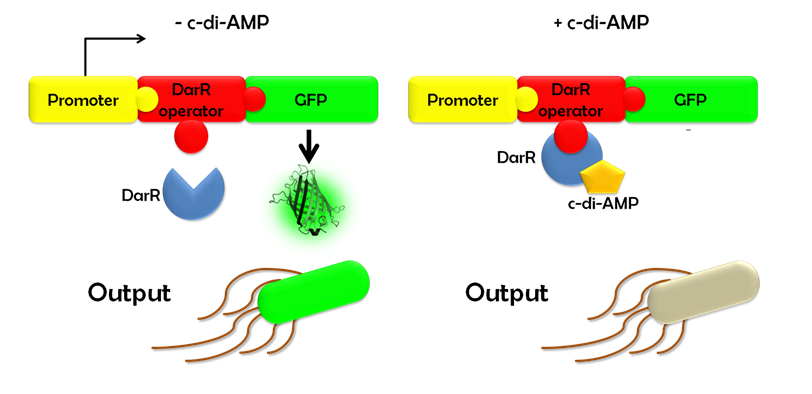
Ideally, this c-di-AMP-sensing system works in the following way (Fig. 1.3): Without c-di-AMP, DarR is not bound to the DarR operator, so GFP is expressed resulting in green fluorescing cells. With c-di-AMP, DarR can bind to its binding sequence, repressing gfp transcription, leading to non-fluorescent cells. In the same way, the system might react to compounds similar to c-di-AMP. It can therefore be used to screen for compounds similar to c-di-AMP. It can be used in a high-throughput scale, as well. This way, the DarR reporter system will make it possible to find inhibitors and competitors which might be used as antibiotics.
Fig. 1.1. The DarR reporter system was assembled step by step in pSB1C3. If a certain biobrick of a specific cloning step was used as a vector, insert or derived from hybridization oligos or PCR is indicated in blue. The part numbers of the intermediates are given in black. The orientation of the arrows corresponds to the orientation of the element in the vector.
Figure 1.2: The final DarR reporter system consisted of two expression units in diverging orientation.
Figure 1.3: Without c-di-AMP, E. coli cells expressing the DarR reporter system will emit a green fluorescence signal. With c-di-AMP the will not fluoresce.
Reference:
1. Zhang et al.(2013) DarR, a TetR-like Transcriptional Factor, Is a Cyclic Di-AMP-responsive Repressor in Mycobacterium smegmatis. J. Biol. Chem. 288:3085–3096Riboswitch reporter system ([http://parts.igem.org/Part:BBa_K1045002 part BBa_K1045002]):
The ydaO gene from B. subtilis is regulated by c-di-AMP. This control is mediated by a c-di-AMP-binding riboswitch, according to data of a publication that appeared this month (Nelson et al., 2013). Structurally, a riboswitch is a RNA-sequence. Functionally, it is a simple switch. It has two mutually exclusive states: ON and OFF. Depending on the circumstances, it can only be in one of the states. The ydaO-riboswitch is dependent on c-di-AMP; whenever it is present, the riboswitch will be turned OFF, meaning that the gene controlled by it cannot be read.
The ydaO riboswitch was, in addition to DarR, a perfect fit as a c-di-AMP sensor for our antibiotic detector. Hence, we constructed an alternative reporter system.
Since it was unclear where the exact beginning and ending of the ydaO promoter, the ydaO riboswitch and the ydaO RBS were, we created four different constructs (Fig. 2.; [http://parts.igem.org/Part:BBa_K1045004 BBa_K1045004], [http://parts.igem.org/Part:BBa_K1045005 BBa_K1045005], [http://parts.igem.org/Part:BBa_K1045006 BBa_K1045006], [http://parts.igem.org/Part:BBa_K1045007 BBa_K1045007]). The sequence of [http://parts.igem.org/Part:BBa_K1045004 BBa_K1045004] was cloned in front of the reporter gene CFP ([http://parts.igem.org/Part:BBa_E0020 BBa_E0020]). For the termination of transcription, we exploited the terminator on pSB1C3. In principle, the riboswitch reporter system might respond in exactly the same way to c-di-AMP, as the DarR reporter system does: Without c-di-AMP, E. coli cells harboring [http://parts.igem.org/Part:BBa_K1045002 BBa_K1045002] will fluoresce blue. In contrast, when c-di-AMP is present, the ydaO riboswitch will terminate transcription of cfp leading to non-fluorescent cells (Fig. 2.2). This way, we can scan substances which can act on the riboswitch as c-di-AMP does. This might lead to the identification of competitors or inhibitors for future antibiotics, as well.
Figure 2.1. The riboswitch reporter system is composed of the ydaO riboswitch and cfp as a reporter gene.
Figure 2.2. In theory, E. coli cells transformed with the riboswitch reporter system will fluoresce blue when c-di-AMP is absent. When c-di-AMP is present, however, the ydaO riboswitch will prevent expression of CFP.
Reference:
Peter Y Watson & Martha J Fedor (2012) “The ''ydaO'' motif is an ATP-sensing riboswitch in Bacillus subtilis“, Nature Chemical Biology No. 8, pp. 963-96
Results
The DarR Reporter System
To test the DarR reporter system, E. coli cells were transformed with the DarR reporter system ([http://parts.igem.org/Part:BBa_K1045017 BBa_K1045017]) (Fig. 3, right image) or with a control plasmid containing the GFP expression unit only ([http://parts.igem.org/Part:BBa_K1045013 BBa_K1045013]) (Fig. 3, left image). Both strains were grown without c-di-AMP and subjected to fluorescence microscopy. While the control strain showed green fluorescing cells, the cells of the DarR reporter system strain barely fluoresced, indicating that DarR acts as a strong repressor in E. coli.
Figure 3: To test the DarR reporter system, fluorescence microscopy of E. coli transformed with a control plasmid (left) and E.coli cells harboring the DarR reporter system (right) was done. Both images are merges of GFP channel and bright field.
The Riboswitch Reporter System
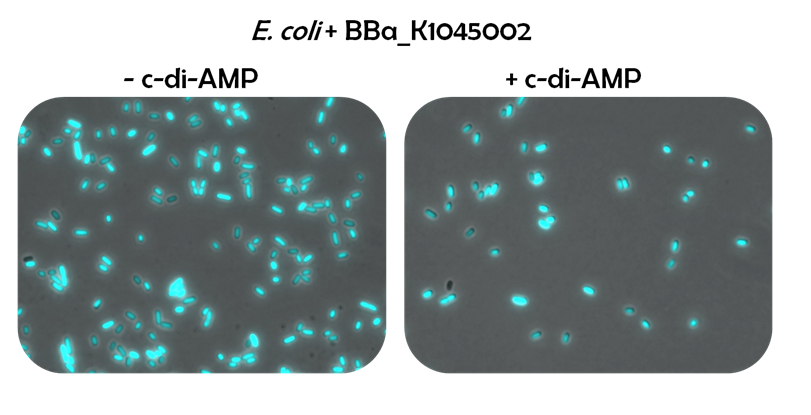
For testing the riboswitch reporter system, E.coli was transformed with [http://parts.igem.org/Part:BBa_K1045002 BBa_K1045002] and grown either with c-di-AMP or without. The cells were analyzed by fluorescence microscopy. A strong fluorescence for both strains could be observed, indicating that the reporter construct is highly expressed from the B. subtilis promoter. However, an influence of c-di-AMP was not seen.
Figure 4: E. coli cells containing the riboswitch reporter system were grown without (left) or with (right) c-di-AMP and studied under the fluorescence microscope.
Discussion
Since we are in urgent need for novel antibiotics, we constructed two different c-di-AMP reporter systems which would allow the screening for compounds affecting c-di-AMP biofunction. One system was based on the c-di-AMP-binding transcriptional repressor DarR. In our system, DarR controlled the expression of the reporter gene GFP. E. coli cells transformed with a vector harboring the DarR reporter system exhibited no green fluorescence compared to cells transformed with a control plasmid lacking the DarR expression unit. This data indicated that DarR seems to be expressed and active as a repressor in E. coli. Even in the absence of c-di-AMP, DarR was likely to bind the DarR operator strongly and repressed GFP expression almost completely. Under these conditions, an intermediate GFP expression level would be more suitable for our purpose to screen for novel antibiotics. Such an intermediate expression level of GFP in the absence of c-di-AMP might be reached in two ways: For instance, one could try a promoter for DarR even weaker than the current one. A more successful approach might be directed mutagenesis of the DarR operator to reduce the binding affinity of DarR to this DNA sequence.
Regarding the alternative reporter system involving the c-di-AMP-responsive ydaO riboswitch, optimization is needed, as well. The results obtained from fluorescence microscopy showed that the riboswitch reporter construct is expressed in E. coli though the native Bacillus promoter and Bacillus RBS were employed. Hence, E. coli seems to be able to use these elements. The strong CFP fluorescence of the E. coli cells even suggests a strong promoter activity. The high transcript levels that might be caused by this strong promoter could account for the observed ineffectiveness of exogenously applied c-di-AMP: There might be more riboswitch than c-di-AMP to bind to. Consequently, one could try to increase the c-di-AMP amounts. Alternatively, one could fuse the riboswitch biobrick [http://parts.igem.org/Part:BBa_K1045005 BBa_K1045005] to a weaker promoter to reduce the mRNA levels. Another possibility might be to switch to low-copy plasmids for expression of the reporter system. If despite reduced transcript levels, c-di-AMP has no effect on the riboswitch reporter system, E. coli might be unable to take up c-di-AMP. Yet, this would not put an end to the construction of a screening system for antibiotics targeting c-di-AMP. It has been reported, that B. subtilis is able to take up c-di-AMP (Oppenheimer-Shaaman et al., 2011). Thus, one could put more effort in unraveling the ways by which B. subtilis and other bacteria take up c-di-AMP. This knowledge might allow the engineering of E. coli regarding c-di-AMP uptake. In parallel to this time-consuming approach, one could try to synthesize c-di-AMP in vivo by expressing diadenylate cyclases in E. coli cells containing an optimized c-di-AMP reporter system. Using DACs of different activity, the output of the reporter system could be characterized. Finally, having a well characterized c-di-AMP reporter system combined with a DAC, it is possible to screen in vivo for antibiotics interfering not only with the function of the essential signaling nucleotide c-di-AMP, but also for its essential biosynthesis enzyme, the DAC.
 "
"