Team:Paris Saclay/Notebook/July/3
From 2013.igem.org
Notebook : July 3
Summary:
FNR regulator system:
- Continued what we started yesterday. Observed the Petri dish, selected the colonies. 4 colonies in total, they were 2 include FNR+plasmid in PSB1C3 and 2 from the control.
- Used 4 primers: VF2, VR, Pfnr_up, Pfnr_down for the verification test. They were designed to cut 4 special sites for creating 3 different regions on plasmid chain: VF2/VR, VF2/PFNR_down, PFNR_up/VR. After the amplification, those PCR products had been put on electrophoresis gel for the verification.
Lab work
- A.aero/anaerobic regulation system
- 2.BioBrick RBS+LacZ+terminator in plasmid PSB1C3
- BioBrick RBS+amilCP+terminator in plasmid PSB1C3
Observation
After the incubation overnight, we observed the Petri dishes.
| colonies | Normal concentration | High concentration |
| control | ||
| Fnr in plasmid PSB1C3 |
We picked up 4 colonies for further test (2 include FNR+plasmid in PSB1C3 and 2 from the control)
Primer and PCR
VF2, VR, Pfnr_up, Pfnr_down are 4 primes that we used for plasmid restriction. The primers, if the promoter fnr entre the plasmid successfully will amplify 3 sub-pieces with specific size. They are VF/VR, VF/Pfnr_down, Pfnr_up/VR.

So we had prepared 4(colonies)*3(amplification) = 12 PCR tubes.
- Dream Taq(5µg/µl):2µl
- Buffer (Dream Taq) 10X:10µl
- dNTP:2µl
- Primer (F/R;F/fnr_R;fnr_F/R):2µl+2µl
- H2O:82µl
- Total:100µl (volume for 4 tubes, so 25µl for each)
PCR programe
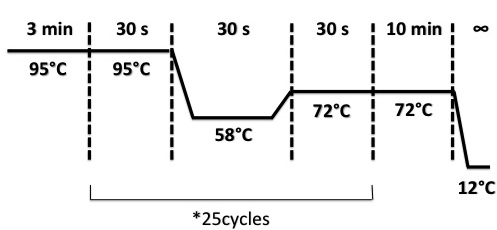
The PCR products was put on a gel of agarose 1.5% with BET (1.5%), migrated during 30 min at 100V. the picture analysis was be delayed to the next day.
Culture confirmation
We performed another colonies seeding. 2 colonies selected from each Petri dish were seeded in liquid medium with their corresponding antibiotic at 37°C under stirring during 1 night.
| Previous day | Back to calendar | Next day |
 "
"