From 2013.igem.org
Aug 19
Colony PCR
Kojima
| Sample | base pair
|
| 8/18 J23100 control -(1) | 1142
|
| 8/18 J23100-RBS-GFP-DT -(1) | 1156
|
| 8/18 J23100-RBS-GFP-DT -(2) | 1156
|
| 8/18 J23100-RBS-luxR-DT -(1) | 1271
|
| 8/18 J23100-RBS-luxR-DT -(2) | 1271
|
| 8/18 J23100-RBS-lacZα-DT -(1) | 670
|
| 8/18 J23100-RBS-lacZα-DT -(2) | 670
|
| 8/18 RBS-lysis3 -(1) | 997
|
| 8/18 RBS-lysis3 -(2) | 997
|
| 8/18 P0440(RBS-tetR-DT) -(1) | 1154
|
| 8/18 P0440(RBS-tetR-DT) -(2) | 1154
|
| negative control | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min | 30cycles
|
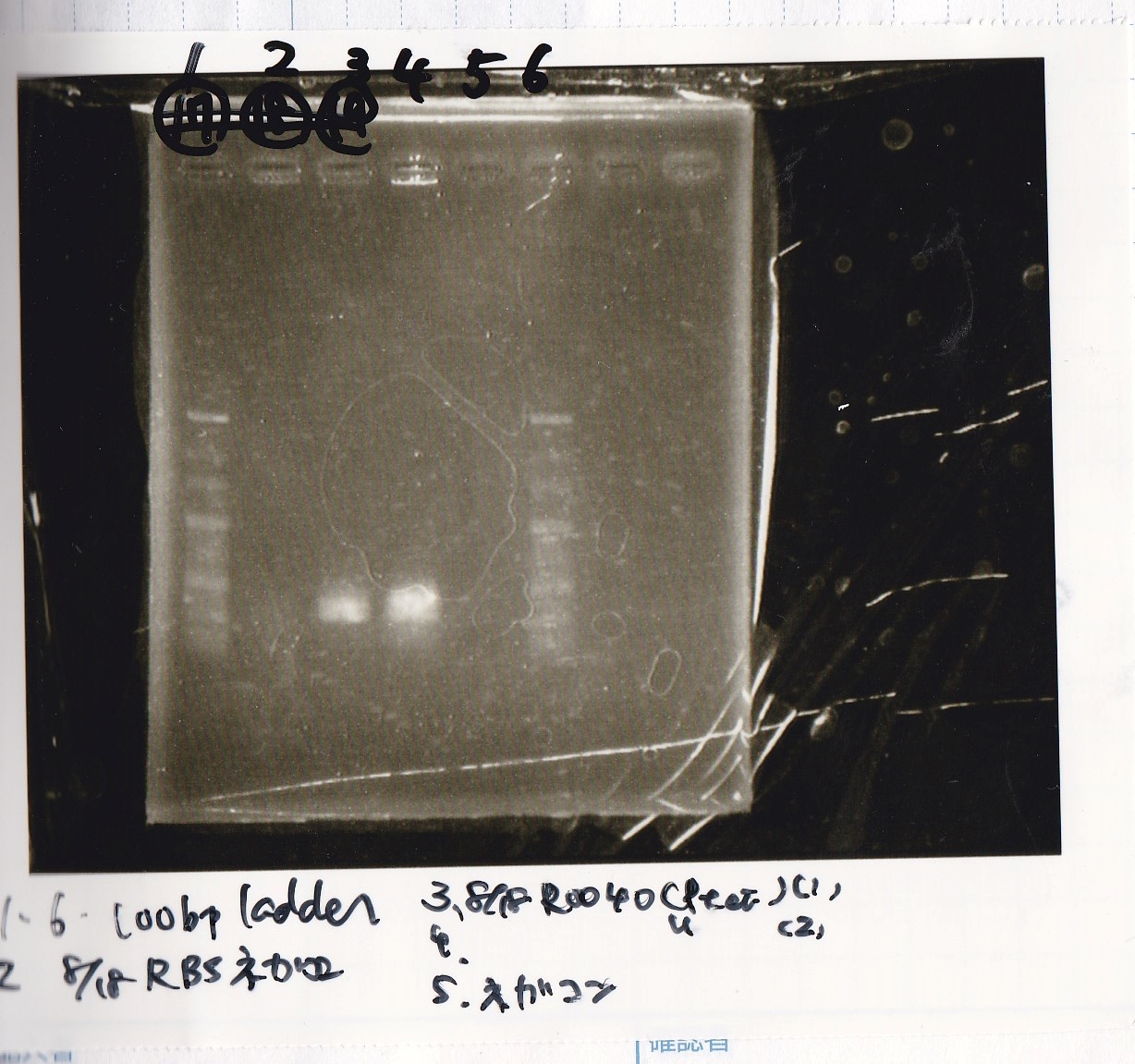
| Sample | base pair
|
| 8/18 RBS control -(1) | --
|
| 8/18 R0040(Ptet) -(1) | 686
|
| 8/18 R0040(Ptet) -(2) | 686
|
| negative control | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
File:Igku Aug19electrophoresis2.png
Ashida
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 3 | 8/18 K117000(lysis2) -1 | XbaI & PstI
|
| 5 | 8/18 K117000(lysis2) -1 | XbaI & PstI
|
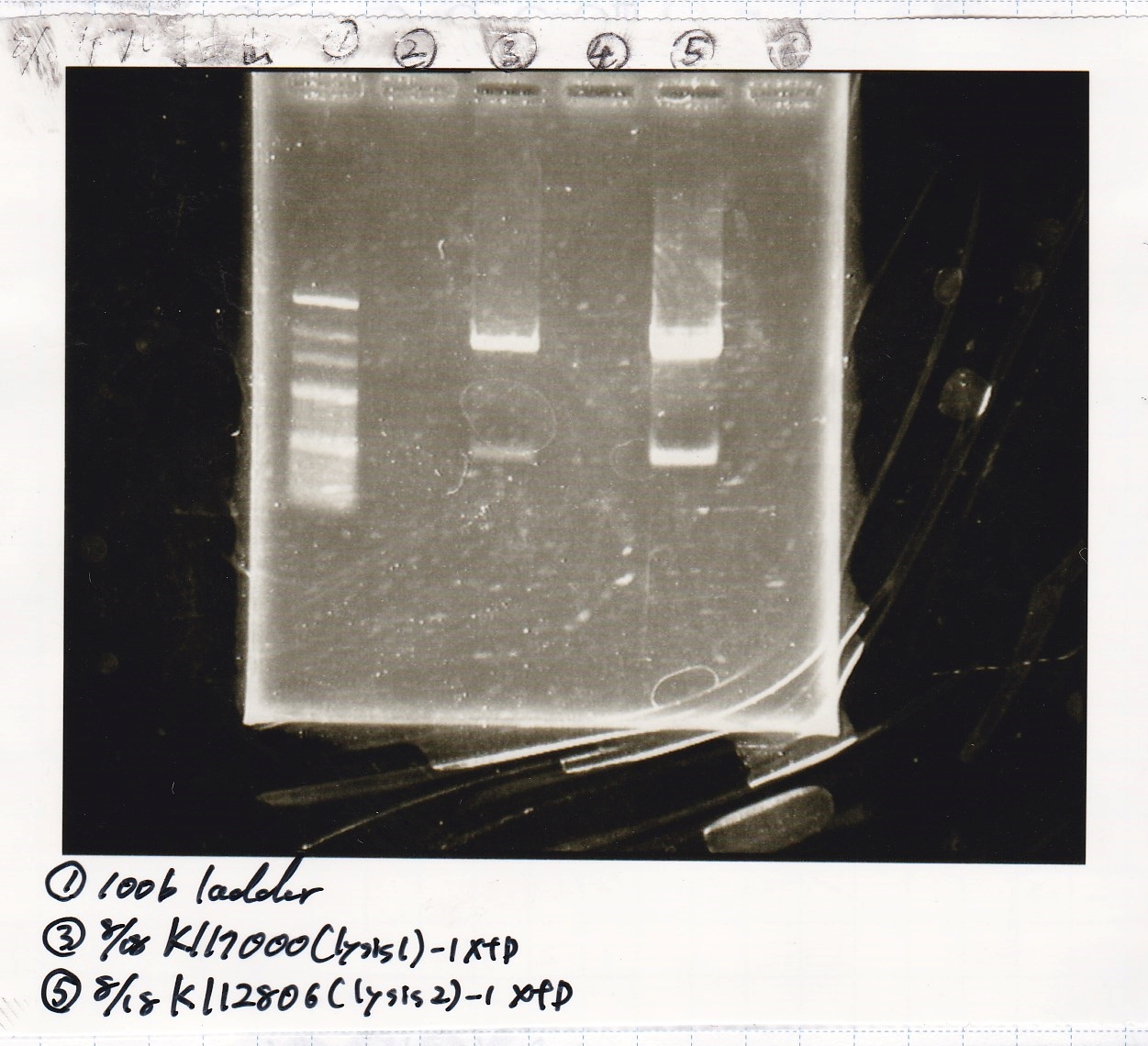
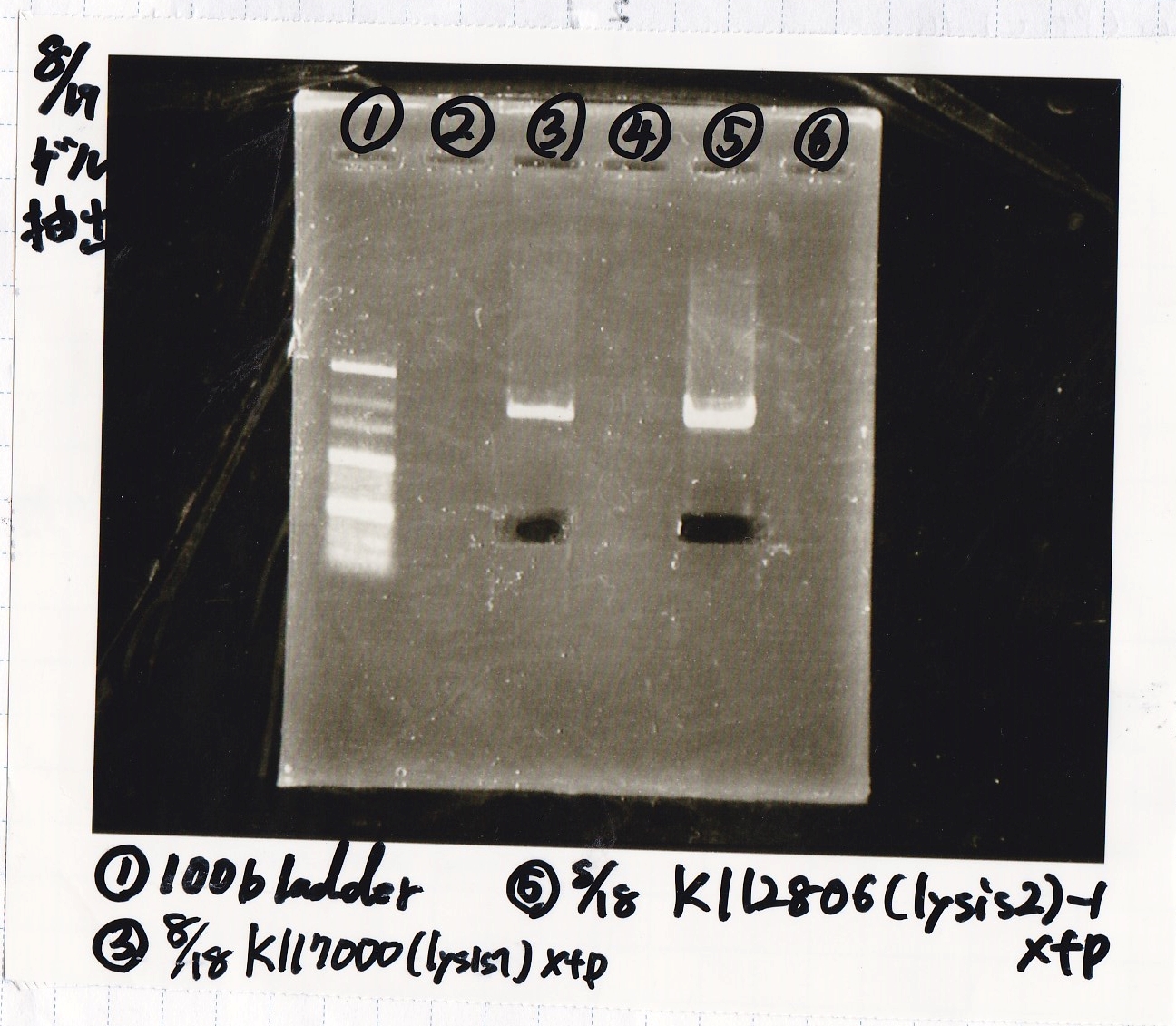
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/18 lysis2-1(XbaI & PstI) | 3.1 | 3.64 | 0.19
|
| 8/18 lysis2-1(XbaI & PstI) | 3.4 | 1.89 | 0.13
|
LB Medium Plate
Nakamoto
| volume | 200ml
|
| Bacto T2ypton | 2g
|
| Bacto yeast extract | 1g
|
| NaCl | 1g
|
| Agar Pouder | 2g
|
Restriction Enzyme Digestion
Tatsui
| | lysis2-1 | XbaI | PstI | 10xbuffer | BSA | MilliQ | total
|
| 2 cuts | 10µL | 1µL | 1µL | 3µL | 3µL | 12µL | 30µL
|
| 1 cut | 2µL | 0.2µL | -- | 1µL | 1µL | 5.8µL | 10µL
|
| 1 cut | 2µL | -- | 0.2µL | 1µL | 1µL | 5.8µL | 10µL
|
| NC | 2µL | -- | -- | 1µL | 1µ | 6µL | 10µL
|
Murata and Okazaki
| | 8/19 lysis1-1 | XbaI | PstI | 10xbuffer | BSA | MilliQ | total
|
| 2 cuts | 2µL | 0.5µL | 0.8µL | 2µL | 2µL | 13µL | 20µL
|
| 1 cut | 0.5µL | 0.2µL | -- | 1µL | 1µL | 7.3µL | 10µL
|
| 1 cut | 0.5µL | -- | 0.2µL | 1µL | 1µL | 7.3µL | 10µL
|
| NC | 0.5µL | -- | -- | 1µL | 1µL | 7.5µL | 10µL
|
Ligation
No name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | RBS | 0.3 | lysis2 | 7.4 | 4
|
| NC | RBS | 0.3 | MilliQ | 7.4 | 4
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | 8/19 lysis1 | XbaI | PstI
|
| 3 | 8/19 lysis1 | XbaI | --
|
| 4 | 8/19 lysis1 | -- | PstI
|
| 5 | 8/19 lysis1 | -- | --
|
| 6 | 8/19 lysis1(Gel Extraction Product) | XbaI | PstI
|
Transformation
Murata and Kojima
| Name | Sample | Competent Cells | Plate
|
| 8/18 Plux+RBS-GFP-DT | 2µL | 20µL | CP
|
| 8/18 Plux(NC) | 2µL | 20µL | CP
|
| 8/19 RBS+lysis2 | 2µL | 20µL | Amp
|
| 8/15 Pbad-araC | 2µL | 20µL | Kan
|
No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | 8/19 lysis1 ① | XbaI & PstI
|
fig before
fig after
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/19 lysis1 ① (XbaI & PstI) | 87.8 | 1.06 | 0.82
|
Liquid Culture
Ashida
| Sample | medium
|
| 8/18 Ptet -1 | Plusgrow medium(+CP)
|
| 8/18 Ptet -2 | Plusgrow medium(+CP)
|
| 8/18 RBS-tetR-DT -1 | Plusgrow medium(+CP)
|
| 8/18 RBS-tetR-DT -2 | Plusgrow medium(+CP)
|
| 8/18 J23100-GFP -1 | Plusgrow medium(+Amp)
|
| 8/18 J23100-luxR -1 | Plusgrow medium(+Amp)
|
| 8/18 J23100-luxR -2 | Plusgrow medium(+Amp)
|
| 8/18 J23100-lacZα -1 | Plusgrow medium(+Amp)
|
| 8/18 RBS-lysis3 -1 | Plusgrow medium(+Amp)
|
| 8/18 RBS-lysis3 -2 | Plusgrow medium(+Amp)
|
Master Plate
Ashida
| Number | Use LB plate(+Amp)
|
| 1 | 8/18 J23100-GFP -1
|
| 2 | 8/18 J23100-luxR -1
|
| 3 | 8/18 J23100-luxR -2
|
| 4 | 8/18 J23100-lacZα -1
|
| 5 | 8/18 RBS-lysis3 -1
|
| 6 | 8/18 RBS-lysis3 -2
|
| Number | Use LB plate(+CP)
|
| 1 | 8/18 Ptet -1
|
| 1 | 8/18 Ptet -2
|
| 3 | 8/18 RBS-tetR-DT -1
|
| 4 | 8/18 RBS-tetR-DT -2
|
 "
"


