| 5 | }
File:Igku xxxxxx.xxx
Ligation
じっけんしゃ
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | name | xx µL | name | xx µL | xx µL
|
| NC | name | xx µL | name | xx µL | xx µL
|
Colony PCR
No name
| PreDenature | Denature | Annealing | Extension | cycle
|
| °C | °C | °C | °C | --
|
| (◎Д◎) | | | |
|
PCR
No name
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | SasA_fwd primer | SasA_rev primer | MilliQ | total
|
| | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| °C | °C | °C | °C | --
|
| (◎Д◎) | | | |
|
Liquid Culture
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| (・▽・) | | |
|
Ethanol Precipitation
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| | | |
|
Master Plate
No name
| Number | Use LB plate (+抗生物質)
|
| ʅ(´◔౪◔)ʃ |
|
Medium Plate
Aug 8
Transformation
Inoue and Nakamoto
| Name | Sample | Competent Cells | Total | Plate
|
| BBa_J44000 | 2 µL | 20 µL (7/30) | 22 µL | LB (+Amp)
|
| BBa_J44000 | 2 µL | 20 µL (XL10-gold) | 22 µL | LB (+Amp)
|
| BBa_K747028 | 2 µL | 20 µL (XL10-gold) | 22 µL | LB (+CP)
|
| BBa_K747028 | 2 µL | 20 µL (7/30) | 22 µL | LB (+CP)
|
| BBa_J23100 | 2 µL | 20 µL (XL10-gold) | 22 µL | LB (+Amp)
|
| BBa_J23106 | 2 µL | 20 µL (XL10-gold) | 22 µL | LB (+Amp)
|
| BBa_E2030 | 2 µL | 20 µL (XL10-gold) | 22 µL | LB (+Kan)
|
Aug 9
Transformation
Kojima and Nakamoto
| Name | Sample | Competent Cells | Total | Plate
|
| BBa_J44000 | 2 µL | 20 µL | 22 µL | LB (+Amp)
|
| BBa_K747028 | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| BBa_J23100 | 2 µL | 20 µL | 22 µL | LB (+Amp)
|
| BBa_J23106 | 2 µL | 20 µL | 22 µL | LB (+Amp)
|
| BBa_E2030 | 2 µL | 20 µL | 22 µL | LB (+Kan)
|
Aug 10
Colony PCR
Nakamoto
| Sample
|
| 8/9 BBa_J44000 -(1)
|
| 8/9 BBa_J44000 -(2)
|
| NC
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 94 °C | 55 °C | 68 °C | --
|
| 5 min | 30 sec | 30 sec | 30 sec | 30 cycles
|
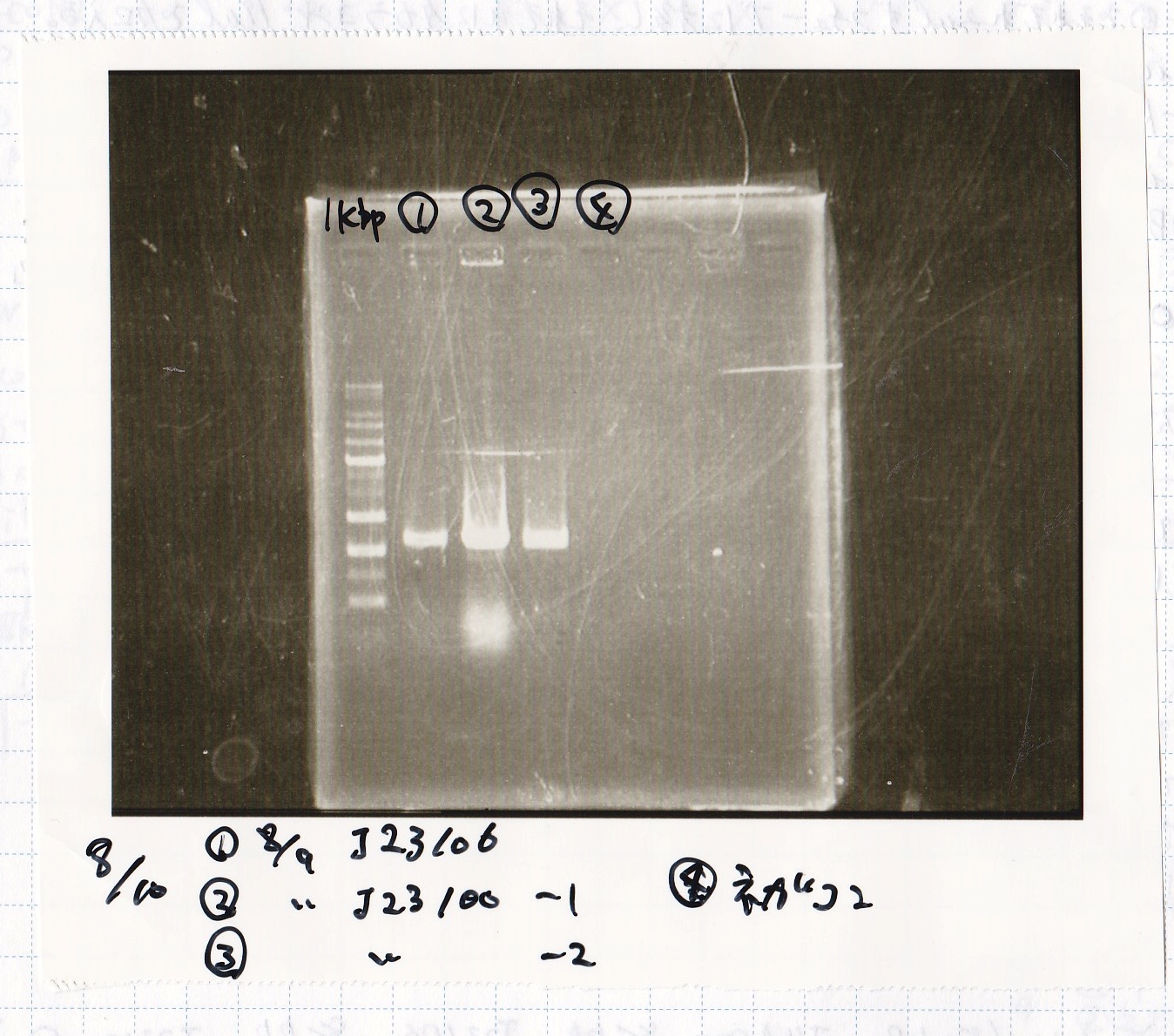
| Sample
|
| 8/9 BBa_J23106
|
| 8/9 BBa_J23100 -(1)
|
| 8/9 BBa_J23100 -(2)
|
| NC
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 94 °C | 55 °C | 68 °C | --
|
| 5 min | 30 sec | 30 sec | 1 min | 30 cycles
|
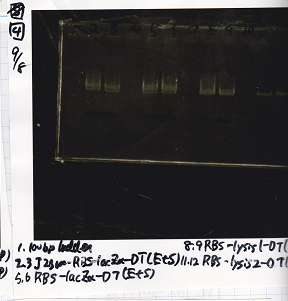
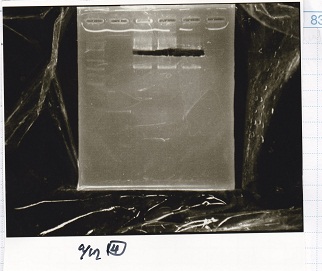
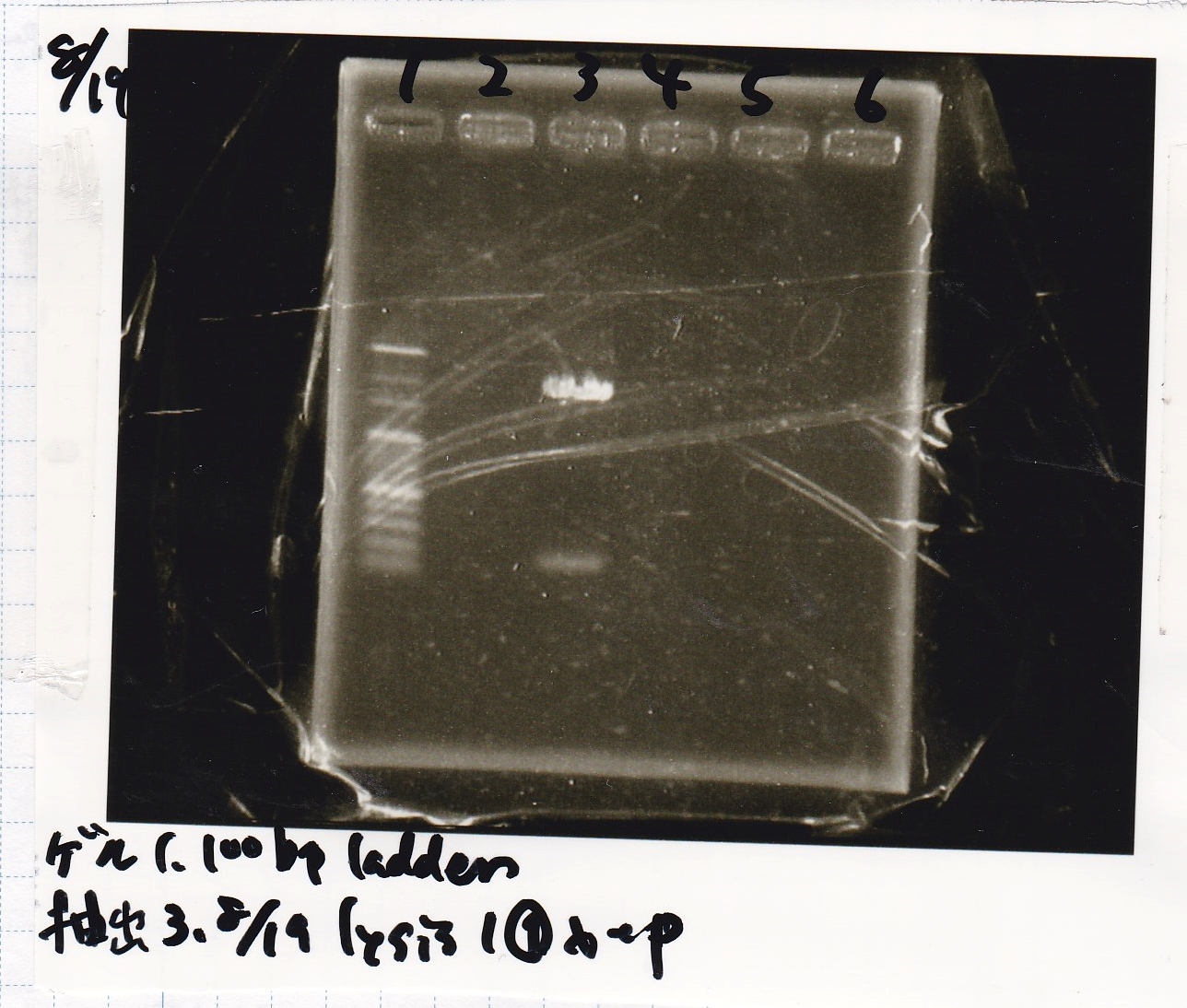
Electrophoresis
Tatsui and Nakamoto
| Lane | Sample
|
| | 100bp ladder
|
| 4 | 8/9 BBa_J44000 -1
|
| 5 | 8/9 BBa_J44000 -2
|
| 6 | NC
|

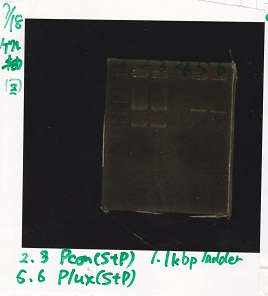
| Lane | Sample
|
| | 1kbp ladder
|
| 1 | 8/9 BBa_J23106
|
| 2 | 8/9 BBa_J23100 -1
|
| 3 | 8/9 BBa_J23100 -2
|
| 4 | NC
|
Liquid Culture
Nakamoto
| Sample | medium
|
| 8/9 BBa-J23106 | LB(+Amp)
|
| 8/9 BBa-J23100-1 | LB(+Amp)
|
| 8/9 BBa-J23100-2 | LB(+Amp)
|
| 8/9 BBa-J44000-1 | LB(+Amp)
|
| 8/9 BBa-J44000-2 | LB(+Amp)
|
LB Medium Plate(+Ampicillin)
Nakamoto & Tatsui
| volume | 200ml
|
| Bacto T2ypton | 2g
|
| Bacto yeast extract | 1g
|
| NaCl | 1g
|
| 0.4M NaOHaq | 500µL
|
| Agarose Pouder | 2g
|
| Ampicillin | 40µL
|
Transformation
Hirano
| Name | Sample | Competent Cells | Total | Plate
|
| tetR | 3µL | 19µL | 22µL | Kan
|
| RBS | 3µL | 19µL | 22µL | Amp
|
| lacZα | 3µL | 19µL | 22µL | CP
|
| lacI | 3µL | 19µL | 22µL | CP
|
| Plac | 3µL | 19µL | 22µL | CP
|
Aug 11
Miniprep
Nakamoto
| DNA | concentration[µg/mL]
|
| 8/9 BBa_J44000 -(2) | 100
|
| 8/9 BBa_J23106 | 154
|
| 8/9 BBa_J23100 | 144
|
| 8/10 T7-His-FT | 92
|
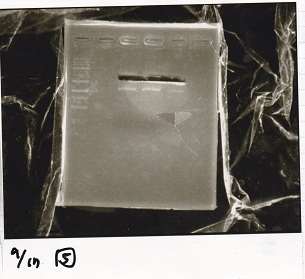
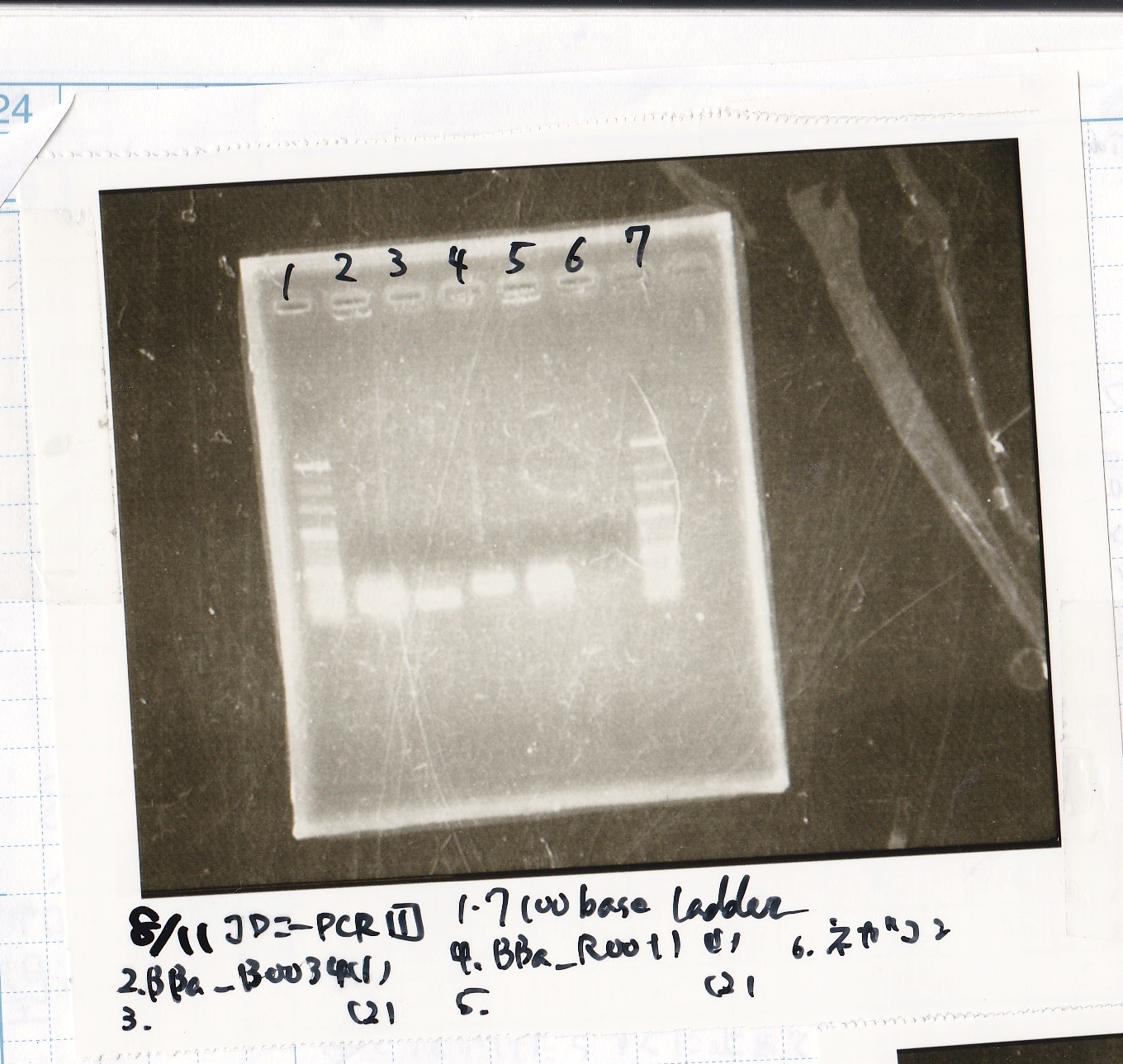
Colony PCR
Kojima and Yasuda
| Sample
|
| BBa_B0034 -(2)
|
| BBa_B0034 -(1)
|
| BBa_R0011 -(2)
|
| BBa_R0011 -(1)
|
| NC
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 94 °C | 55 °C | 68 °C | --
|
| 5 min | 30 s | 30 s | 20 s | 30 cycles
|
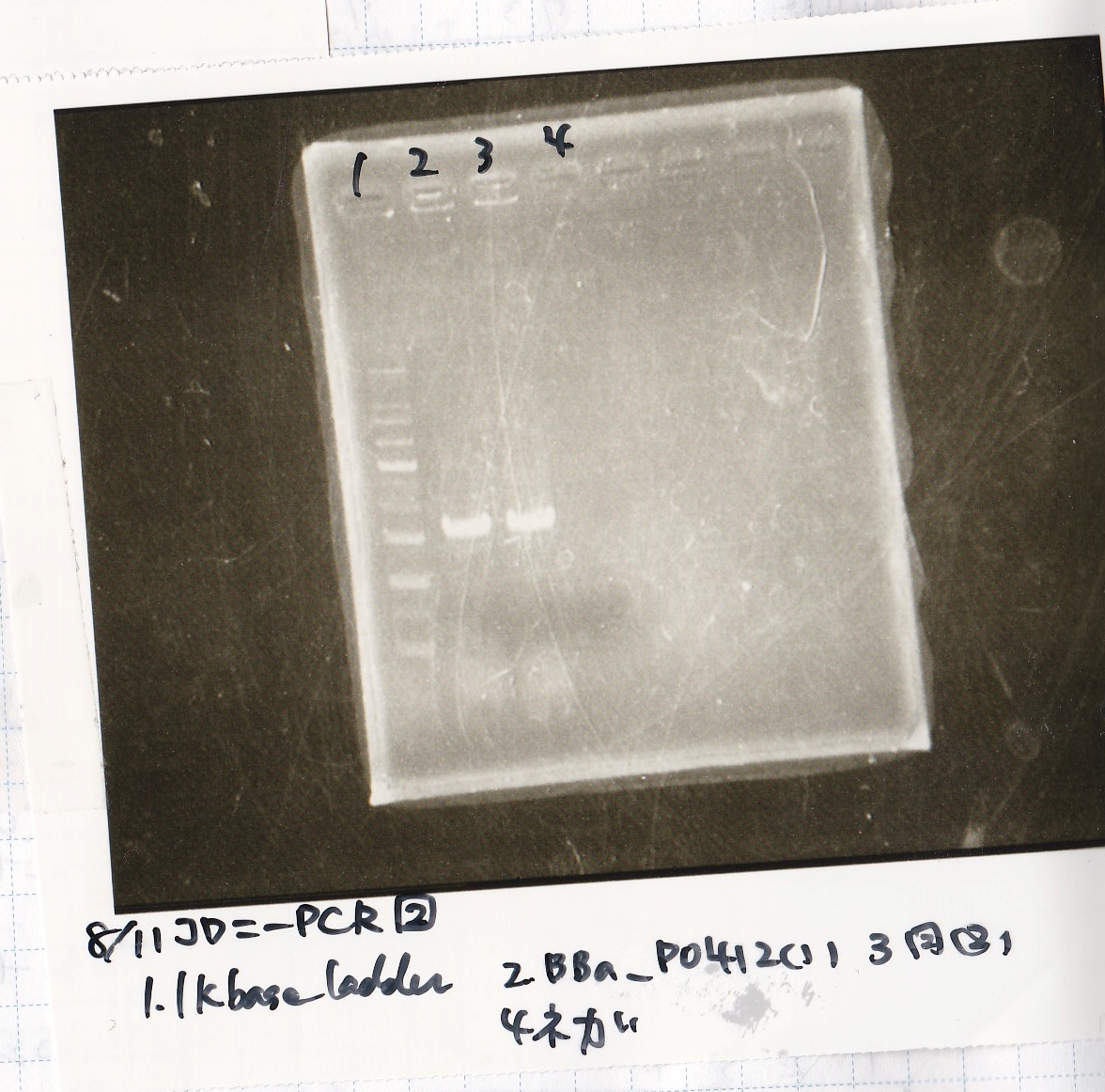
| Sample
|
| BBa_D0412 -(1)
|
| BBa_D0412 -(2)
|
| NC
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 94 °C | 55 °C | 68 °C | --
|
| 5 min | 30 s | 30 s | 1 min 30 s | 30 cycles
|
Restriction Enzyme Digestion
Kojima anda Yasuda
| 8/9 BBa_J44000 2 | EcoRI | PstI | 10x buffer H | MilliQ | total
|
| 2 | 0.2 | 0.2 | 1 | 6.6 | 10
|
| 2 | 0.2 | -- | 1 | 6.8 | 10
|
| 2 | -- | 0.2 | 1 | 6.8 | 10
|
| 2 | -- | -- | 1 | 7.0 | 10
|
| 8/9 BBa_J23106 | EcoRI | PstI | 10x buffer H | MilliQ | total
|
| 1.3 | 0.2 | 0.2 | 1 | 7.3 | 10
|
| 1.3 | 0.2 | -- | 1 | 7.5 | 10
|
| 1.3 | -- | 0.2 | 1 | 7.5 | 10
|
| 1.3 | -- | -- | 1 | 7.7 | 10
|
| 8/9 BBa_J23100 2 | EcoRI | PstI | 10x buffer H | MilliQ | total
|
| 1.3 | 0.2 | 0.2 | 1 | 7.3 | 10
|
| 1.3 | 0.2 | -- | 1 | 7.5 | 10
|
| 1.3 | -- | 0.2 | 1 | 7.5 | 10
|
| 1.3 | -- | -- | 1 | 7.7 | 10
|
- incubated at 37 °C for 1 hour.
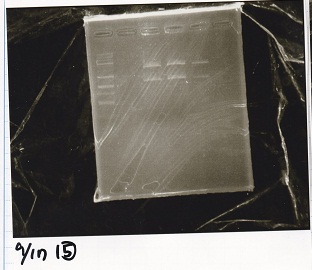
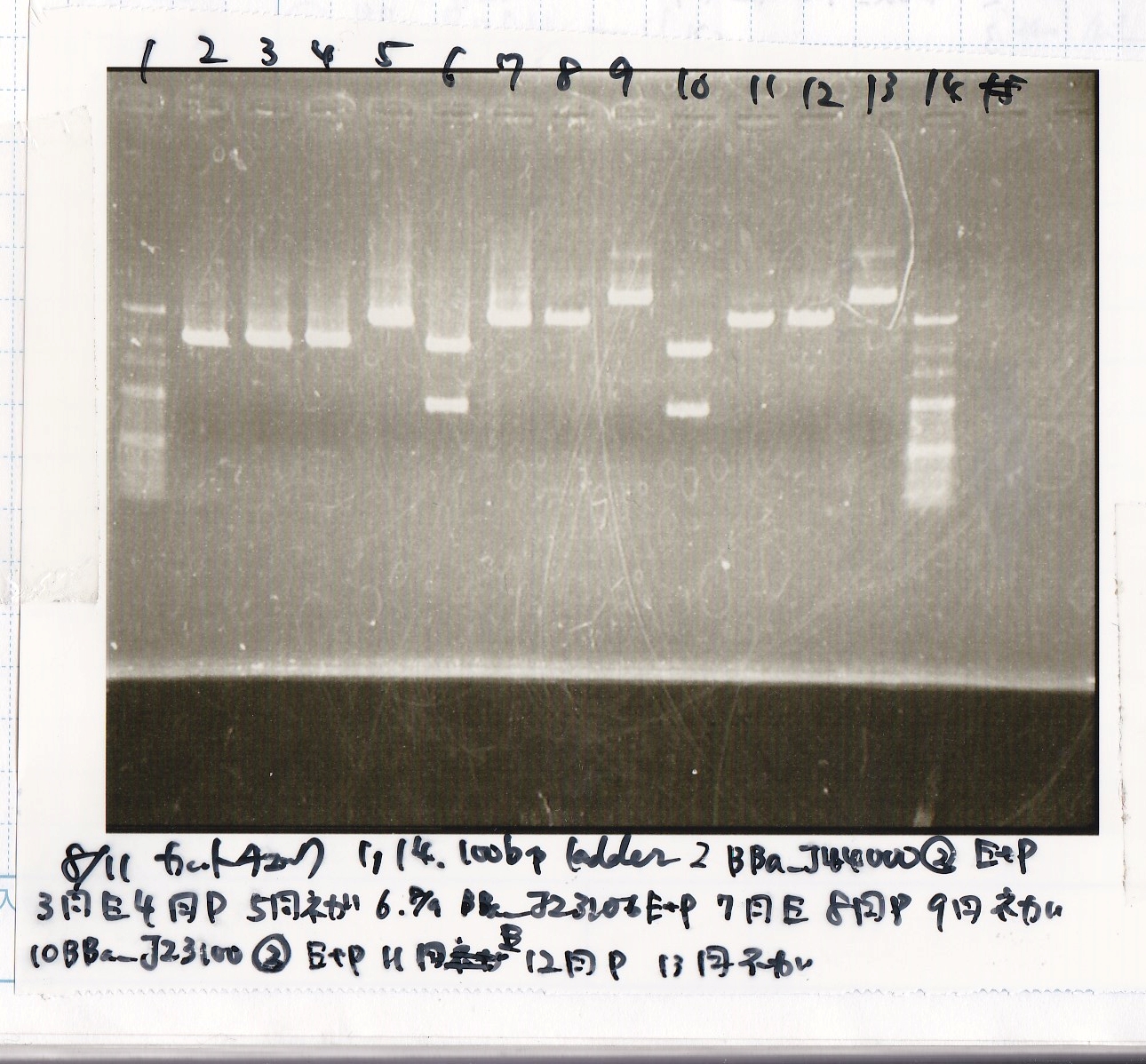
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | 8/9 BBa_J44000 | EcoRI | PstI
|
| 3 | 8/9 BBa_J44000 | EcoRI | --
|
| 4 | 8/9 BBa_J44000 | -- | PstI
|
| 5 | 8/9 BBa_J44000 | -- | --
|
| 6 | 8/9 BBa_J23106 | EcoRI | PstI
|
| 7 | 8/9 BBa_J23106 | EcoRI | --
|
| 8 | 8/9 BBa_J23106 | -- | PstI
|
| 9 | 8/9 BBa_J23106 | -- | --
|
| 10 | 8/9 BBa_J23100 | EcoRI | PstI
|
| 11 | 8/9 BBa_J23100 | EcoRI | --
|
| 12 | 8/9 BBa_J23100 | -- | PstI
|
| 13 | 8/9 BBa_J23100 | -- | --
|
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | BBa_B0034 (1)
|
| 3 | BBa_B0034 (2)
|
| 4 | BBa_R0011 (1)
|
| 5 | BBa_R0011 (2)
|
| 6 | NC
|
| 7 | 100bp ladder
|
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | BBa_P0412 (1)
|
| 3 | BBa_P0412 (2)
|
| 4 | NC
|
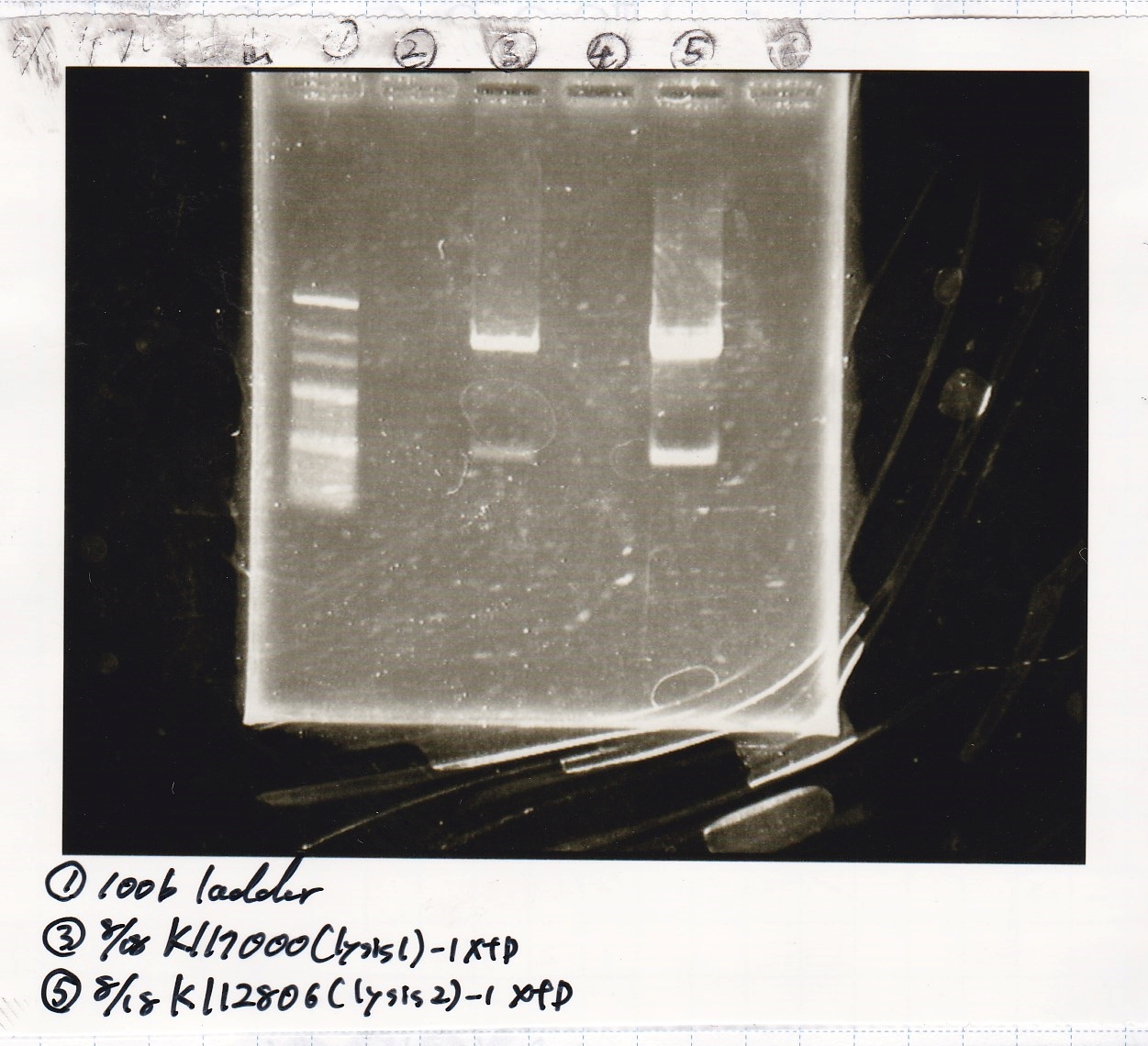
Aug 15
Liquid Culture
Nakamoto
| Sample | medium
|
| 8/10 BBa_B0034(1) | LB (+Amp)
|
| 8/10 BBa_B0034(2) | LB (+Amp)
|
| 8/10 BBa_R0011(1) | LB (+CP)
|
| 8/10 BBa_R0011(2) | LB (+CP)
|
| 8/10 BBa_P0412(1) | LB (+CP)
|
| 8/10 BBa_P0412(2) | LB (+CP)
|
Transformation
Nakamoto
| Name | Sample | Competent Cells | Total | Plate
|
| BBa_I0500(Pbad/araC) | 2 | 17 | 19 | LB (+Kan)
|
| BBa_F1610(RBS-luxI-DT) | 2 | 17 | 19 | LB (+Amp)
|
| BBa_R0061(Plux) | 2 | 17 | 19 | LB (+CP)
|
| BBa_I13504(RBS-GFP-DT) | 2 | 17 | 19 | LB (+CP)
|
| BBa_I0462(RBS-luxR-DT) | 2 | 17 | 19 | LB (+Amp)
|
| BBa_K117000(lysis1) | 2 | 17 | 19 | LB (+CP)
|
| BBa_K112806(lysis2) | 2 | 17 | 19 | LB (+CP)
|
| BBa_K748002(lysis3) | 2 | 17 | 19 | LB (+CP)
|
| BBa_B0015(DT) | 2 | 17 | 19 | LB (+CP)
|
| BBa_Q04400(RBS-tetR-DT-Ptet) | 2 | 17 | 19 | LB (+Kan)
|
| BBa_I732020(RBS-lacZα-DT) | 2 | 17 | 19 | LB (+CP)
|
| BBa_J23100 | 2 | 17 | 19 | LB (+Amp)
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| BBa_B0034(1) | 84.1 | 2.11 | 2.01
|
| BBa_B0034(2) | 86.9 | 2.03 | 2.02
|
| BBa_R0011(1) | 78.3 | 2.26 | 2.02
|
| BBa_R0011(2) | 88.9 | 2.24 | 2.02
|
| BBa_P0412(1) | 15.61 | 2.10 | 1.93
|
| BBa_P0412(2) | 12.93 | 2.18 | 1.91
|
Aug 16
Colony PCR
Tatsui and Ashida
| Sample | base pair
|
| 8/15 BBa_I13504(RBS-GFP-DT)(1) | 1174
|
| 8/15 BBa_I13504(RBS-GFP-DT)(2) | 1174
|
| 8/15 BBa_I0462(RBS-luxR-DT)(1) | 1174
|
| 8/15 BBa_I0462(RBS-luxR-DT)(2) | 1174
|
| 8/15 BBa_K748002(lysis3) | 1053
|
| NC | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min | 30cycles
|
| Sample | base pair
|
| 8/15 BBa_R0061(Plux) | 344
|
| 8/15 BBa_K117000(lysis1)(1) | 456
|
| 8/15 BBa_K117000(lysis1)(2) | 456
|
| 8/15 BBa_B0015(DT) | 443
|
| 8/15 BBa_I732020(RBS-lacZα-DT) | 703
|
| NC | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 40s | 30cycles
|
Electrophoresis
No name
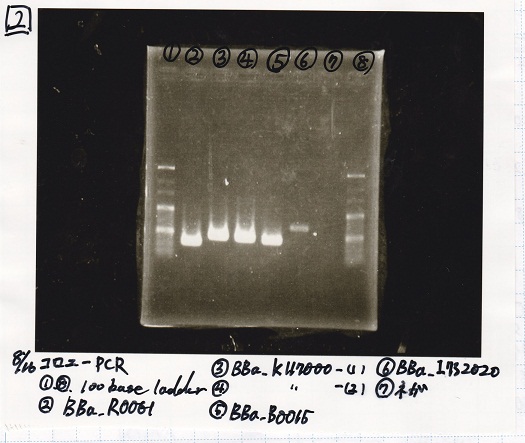
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | BBa_I13504(RBS-GFP-DT) -(1)
|
| 3 | BBa_I13504(RBS-GFP-DT) -(2)
|
| 4 | BBa_I0462(RBS-luxR-DT) -(1)
|
| 5 | BBa_I0462(RBS-luxR-DT) -(2)
|
| 6 | BBa_K748002(lysis3)
|
| 7 | NC
|
| 8 | 100bp ladder
|
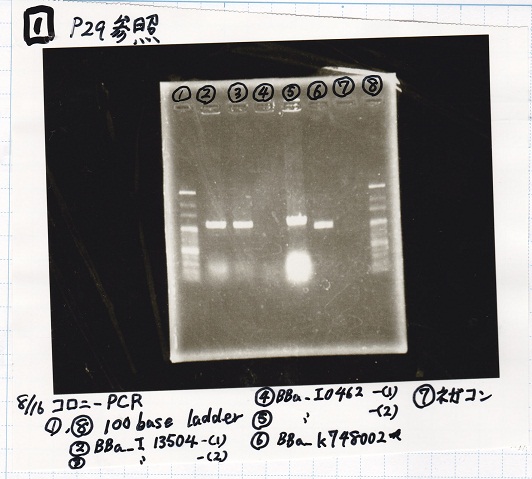
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | BBa_R0061(Plux)
|
| 3 | BBa_K117000(lysis1) -(1)
|
| 4 | BBa_K117000(lysis1) -(1)
|
| 5 | BBa_B0015(DT)
|
| 6 | BBa_I732020(RBS-lacZα-DT)
|
| 7 | NC
|
| 8 | 100bp ladder
|
Master Plate
Tatsui
| Number | Use LB plate (+CP)
|
| 1 | BBa_I13504(RBS-GFP-DT)(1)
|
| 2 | BBa_I13504(RBS-GFP-DT)(2)
|
| 3 | BBa_K748002(lysis3)
|
| 4 | BBa_R0061(Plux)
|
| 5 | BBa_K117000(lysis1)(1)
|
| 6 | BBa_K117000(lysis1)(2)
|
| 7 | BBa_B0015(DT)
|
| 8 | BBa_I732020(RBS-lacZα-DT)
|
| Number | Use LB plate (+Amp)
|
| 1 | BBa_I0462(RBS-luxR-DT)(1)
|
| 2 | BBa_I0462(RBS-luxR-DT)(2)
|
Transformation
Hirano
| Name | Sample | Competent Cells | Total | Plate
|
| pBR322 | 1µL | 100µL(XL-10gold) | 101µL | Amp
|
| pBR322 | 1µL | 50µL(takara) | 51µL | Amp
|
| Pbad/araC | 2µL | 20µL | 22µL | Kan
|
| RBS-luxI-DT | 2µL | 20µL | 22µL | Kan
|
Liquid Culture
Yoshdia
| Sample | medium
|
| 8/15 BBa_I13504(RBS-GFP-DT))(1) | Plusgrow medium (+CP)
|
| 8/15 BBa_I13504(RBS-GFP-DT)(2) | Plusgrow medium (+CP)
|
| 8/15 BBa_I0462(RBS-luxR-DT)(2) | Plusgrow medium (+Amp)
|
| 8/15 BBa_K748002(lysis3) | Plusgrow medium (+CP)
|
| 8/15 BBa_R0061(Plux) | Plusgrow medium (+CP)
|
| 8/15 BBa_K117000(lysis1)(1) | Plusgrow medium (+CP)
|
| 8/15 BBa_K117000(lysis1)(2) | Plusgrow medium (+CP)
|
| 8/15 BBa_B0015(DT) | Plusgrow medium (+CP)
|
| 8/15 BBa_I732020(RBS-lacZα-DT) | Plusgrow medium (+CP)
|
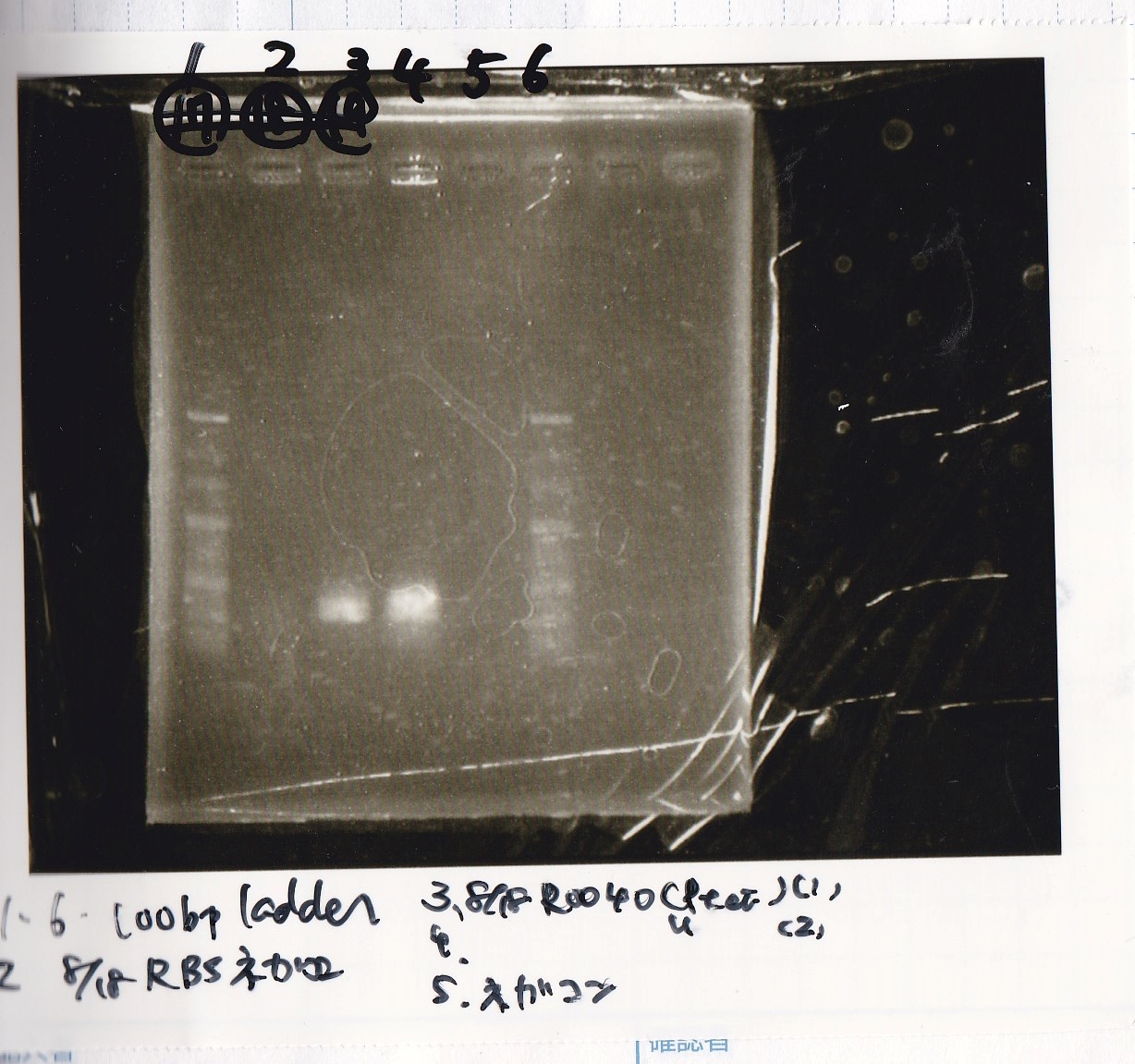
Aug 17
Colony PCR
Nakamoto
| Sample | base pair
|
| 8/15 BBa_K112806(lysis2) -(1) | 828
|
| NC | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min | 30cycles
|
| Sample | base pair
|
| 8/10 BBa_Q0400 -(1) | 3995
|
| NC | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
Electrophoresis
No name
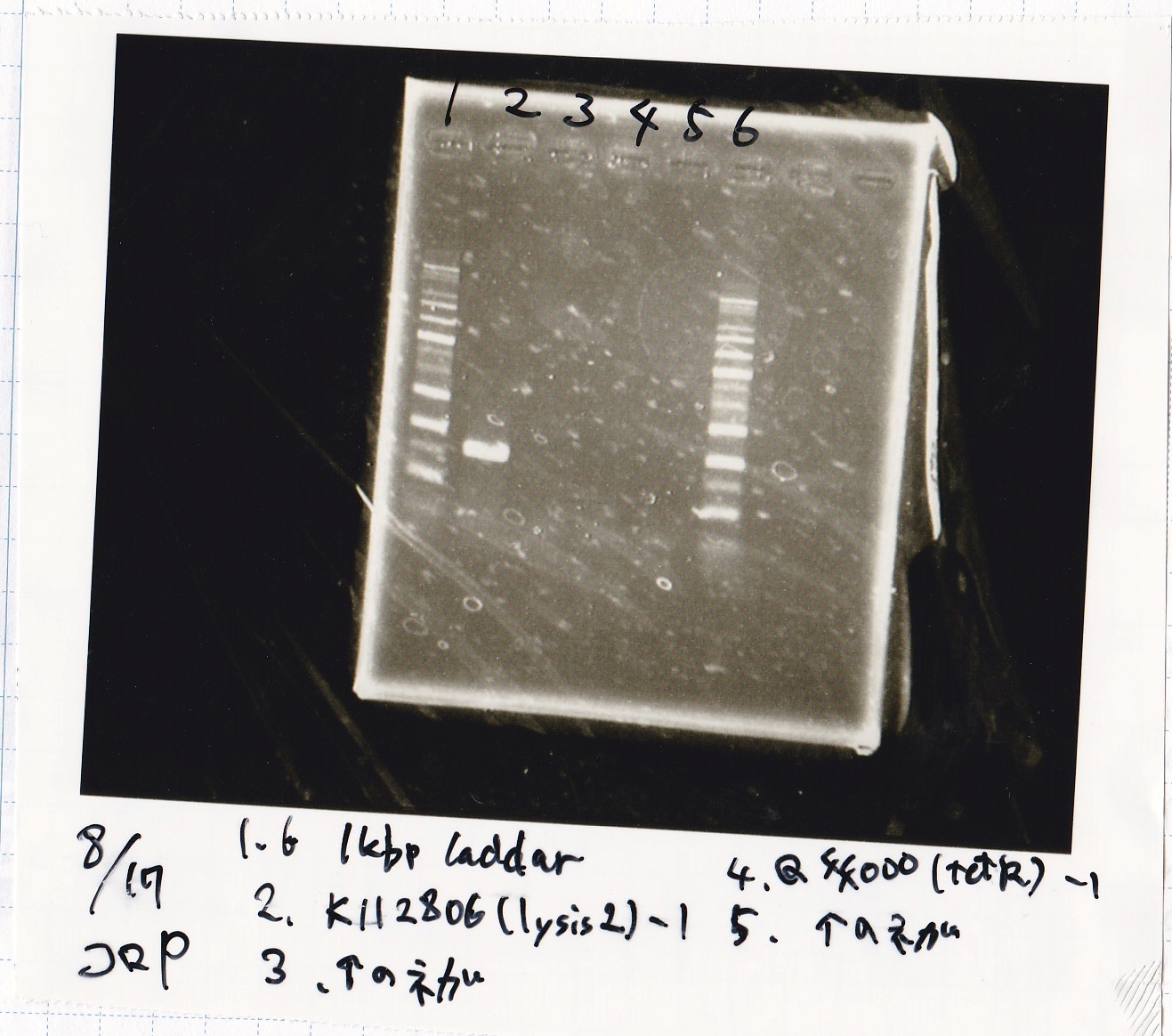
| Lane | Sample
|
| 1 | 1kbp ladder
|
| 2 | K112806(lysis2) -1
|
| 3 | NC
|
| 4 | Q04400(tetR) -1
|
| 5 | NC
|
| 6 | 1kbp ladder
|
Liquid Culture
Nakamoto
| Sample | medium
|
| 8/15 BBa_K112806(lysis2) -(1) | Plusgrow medium(+CP)
|
| 8/15 BBa_Q04400(tetR) -(1) | Plusgrow medium(+Amp)
|
| 8/16 pBR322 -(1) | Plusgrow medium(+Amp)
|
Master Plate
Nakamoto
| Number | Use LB plate (+CP)
|
| 1 | BBa_K112806(lysis2) -(1)
|
| Number | Use LB plate (+Kana)
|
| 1 | BBa_Q04400(tetR) -(1)
|
Miniprep
Nakamoto and Murakami
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| K748002(lysis3) -(1) | 330 | 1.66 | 2.08
|
| B0015(DT) -(1) | 188 | 1.73 | 1.82
|
| R0061(Plux) -(1) | 76 | 1.64 | 1.30
|
| K117000(lysis1) -(1) | 286 | 1.71 | 2.01
|
| K117000(lysis1) -(2) | 248 | 1.69 | 1.78
|
| I732020(RBS-lacZα-DT) -(1) | 198 | 1.71 | 1.93
|
| I13504(RBS-GFP-DT) -(1) | 248 | 1.69 | 2.03
|
| I13504(RBS-GFP-DT) -(2) | 246 | 1.55 | 1.73
|
| I0462(RBS-luxR-DT) -(1) | 326 | 1.65 | 1.74
|
Restriction Enzyme Digestion
Murata, Murakami, Nakamoto, Ashida, Matsumoto and Yoshida
| | 8/11 J23100 -(2) | SpeI | PstI | Buffer | MilliQ | total
|
| 2 cuts | 4.0µL | 1.0µL | 1.0µL | 3.0µL | 21µL | 30µL
|
| 1 cut | 1.3µL | 0.2µL | -- | 1.0µL | 7.5µL | 10µL
|
| 1 cut | 1.3µL | -- | 0.2µL | 1.0µL | 7.5µL | 10µL
|
| NC | 1.3µL | -- | -- | 1.0µL | 7.7µL | 10µL
|
| | 8/15 B0034(RBS) -(2) | SpeI | PstI | Buffer | MilliQ | total
|
| 2 cuts | 6.0µL | 1.0µL | 1.0µL | 3.0µL | 19µL | 30µL
|
| 1 cut | 2.0µL | 0.2µL | -- | 1.0µL | 6.8µL | 10µL
|
| 1 cut | 2.0µL | -- | 0.2µL | 1.0µL | 6.8µL | 10µL
|
| NC | 2.0µL | -- | -- | 1.0µL | 7.0µL | 10µL
|
| | 8/17 K117000(lysis1) -(1) | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 2.0µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 20µL | 30µL
|
| 1 cut | 1.0µL | 0.2µL | -- | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| 1 cut | 1.0µL | -- | 0.2µL | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| NC | 1.0µL | -- | -- | 1.0µL | 1.0µL | 7.0µL | 10µL
|
| | 8/17 K748002(lysis3) -(1) | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 2.0µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 20µL | 30µL
|
| 1 cut | 1.0µL | 0.2µL | -- | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| 1 cut | 1.0µL | -- | 0.2µL | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| NC | 1.0µL | -- | -- | 1.0µL | 1.0µL | 7.0µL | 10µL
|
| | 8/17 I13504(RBS-GFP-DT) -(1) | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 2.0µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 20µL | 30µL
|
| 1 cut | 1.0µL | 0.2µL | -- | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| 1 cut | 1.0µL | -- | 0.2µL | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| NC | 1.0µL | -- | -- | 1.0µL | 1.0µL | 7.0µL | 10µL
|
| | 8/17 I0462(RBS-luxR-DT) -(1) | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 2.0µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 20µL | 30µL
|
| 1 cut | 1.0µL | 0.2µL | -- | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| 1 cut | 1.0µL | -- | 0.2µL | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| NC | 1.0µL | -- | -- | 1.0µL | 1.0µL | 7.0µL | 10µL
|
| | 8/17 I732020(RBS-lacZα-DT) -(1) | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 2.0µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 20µL | 30µL
|
| 1 cut | 1.0µL | 0.2µL | -- | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| 1 cut | 1.0µL | -- | 0.2µL | 1.0µL | 1.0µL | 6.8µL | 10µL
|
| NC | 1.0µL | -- | -- | 1.0µL | 1.0µL | 7.0µL | 10µL
|
| | 8/11 R6061 -(1) | SpeI | PstI | Buffer | MilliQ | total
|
| 2 cuts | 8.0µL | 1.0µL | 1.0µL | 3.0µL | 17µL | 30µL
|
| 1 cut | 3.0µL | 0.2µL | -- | 1.0µL | 5.8µL | 10µL
|
| 1 cut | 3.0µL | -- | 0.2µL | 1.0µL | 5.8µL | 10µL
|
| NC | 3.0µL | -- | -- | 1.0µL | 6.0µL | 10µL
|
- incubate at 37 °C, for 1 h
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | J23100 -② | SpeI | PstI
|
| 2 | J23100 -② | SpeI | --
|
| 3 | J23100 -② | -- | PstI
|
| 4 | J23100 -② | -- | --
|
| 5 | B0034(RBS) -(2) | SpeI | PstI
|
| 6 | B0034(RBS) -(2) | SpeI | --
|
| 7 | B0034(RBS) -(2) | -- | PstI
|
| 8 | B0034(RBS) -(2) | -- | --
|
| 9 | 1kb ladder | -- | --
|
| 10 | K11700(lysis1) -(1) | XbaI | PstI
|
| 11 | K11700(lysis1) -(1) | XbaI | --
|
| 12 | K11700(lysis1) -(1) | -- | PstI
|
| 13 | K11700(lysis1) -(1) | -- | --
|
| 14 | K748002(lysis3) -(1) | XbaI | PstI
|
| 15 | K748002(lysis3) -(1) | XbaI | --
|
| 16 | K748002(lysis3) -(1) | -- | PstI
|
| 17 | K748002(lysis3) -(1) | -- | --
|
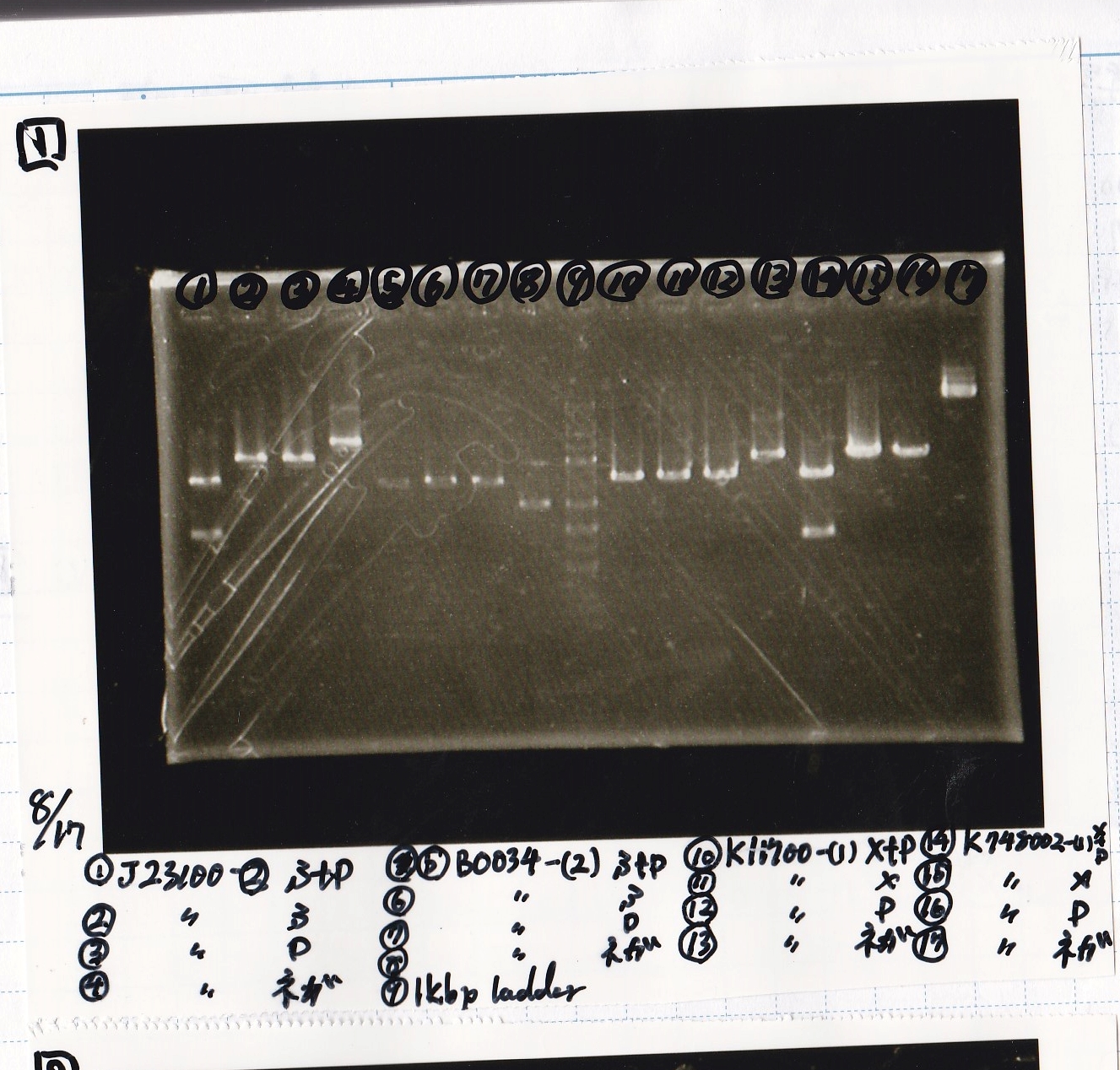
| Lane | Sample | Enzyme1 | Enzyme2
|
| 18 | I13504(RBS-GFP-DT) -(1) | XbaI | PstI
|
| 19 | I13504(RBS-GFP-DT) -(1) | XbaI | --
|
| 20 | I13504(RBS-GFP-DT) -(1) | -- | PstI
|
| 21 | I13504(RBS-GFP-DT) -(1) | -- | --
|
| 22 | I0462(RBS-luxR-DT) -(1) | XbaI | PstI
|
| 23 | I0462(RBS-luxR-DT) -(1) | XbaI | --
|
| 24 | I0462(RBS-luxR-DT) -(1) | -- | PstI
|
| 25 | I0462(RBS-luxR-DT) -(1) | -- | --
|
| 26 | 1kb ladder | -- | --
|
| 27 | I732020(RBS-lacZα-DT) -(1) | XbaI | PstI
|
| 28 | I732020(RBS-lacZα-DT) -(1) | XbaI | --
|
| 29 | I732020(RBS-lacZα-DT) -(1) | -- | PstI
|
| 30 | I732020(RBS-lacZα-DT) -(1) | -- | --
|
| 31 | R0061(Plux) -(1) | SpeI | PstI
|
| 32 | R0061(Plux) -(1) | SpeI | --
|
| 33 | R0061(Plux) -(1) | -- | PstI
|
| 34 | R0061(Plux) -(1) | -- | --
|
Miniprep
Murata
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 8/17 PBR322(TaKaRa) -(1) | 47 | 1.68 | 1.55
|
| 8/17 K112806(lysis2) -(1) | 64 | 1.67 | 1.70
|
Aug 18
Yoshida and Kojima
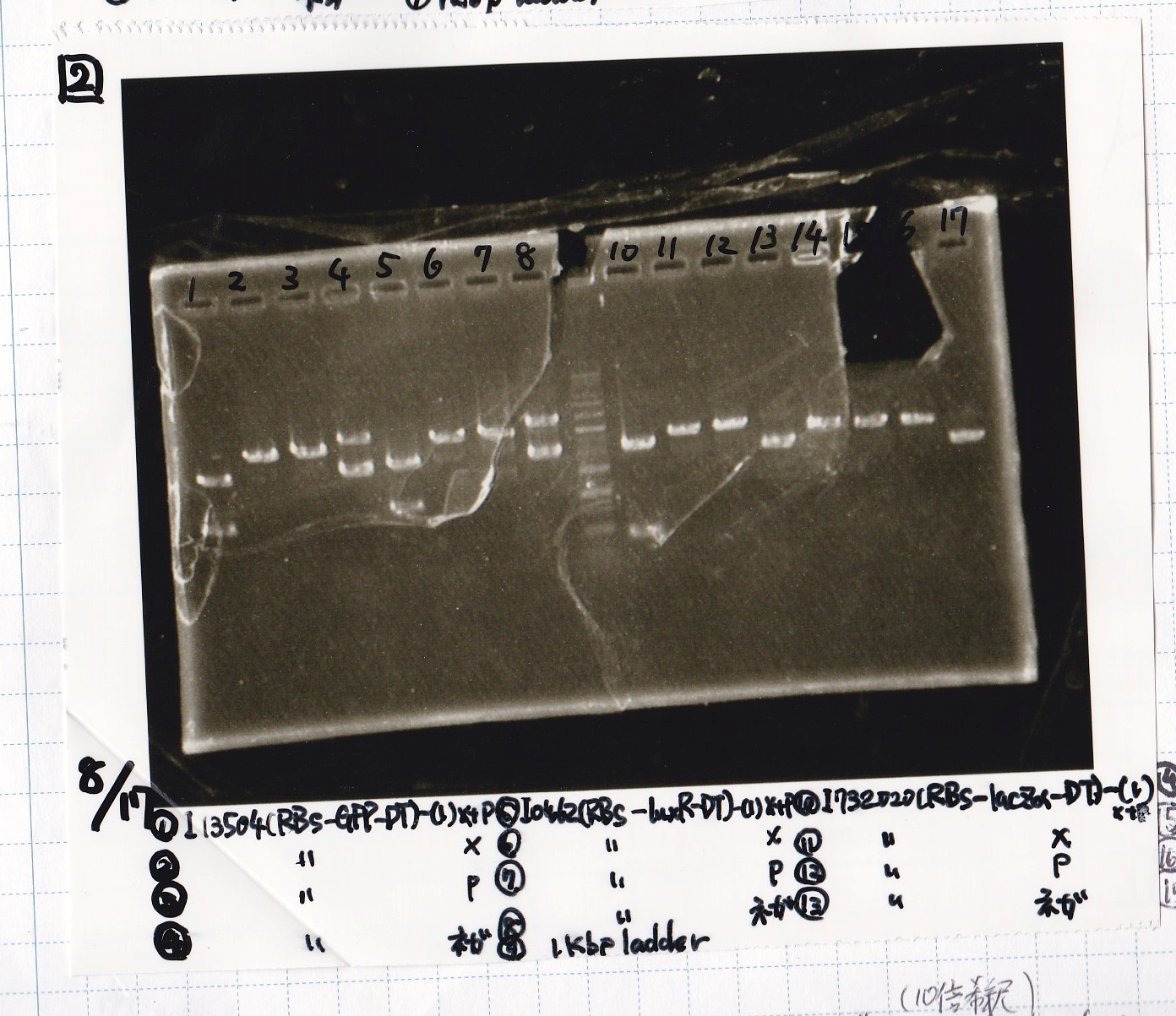
| Lane | DNA | Enzyme
|
| 1 | 100kb ladder | --
|
| 2 | 8/11 J23100 -② | --
|
| 3 | 8/11 J23100 -② | --
|
| 4 | -- | --
|
| 5 | 8/17 K117000(lysis1) -1 | --
|
| 6 | 8/17 K117000(lysis1) -1 | --
|
| 7 | -- | --
|
| 8 | 8/17 K748002(lysis3) -1 | --
|
| 9 | 8/17 K748002(lysis3) -1 | --
|
| 10 | -- | --
|
| 11 | 8/17 I13504(RBS-GFP-DT) -1 | --
|
| 12 | 8/17 I13504(RBS-GFP-DT) -1 | --
|
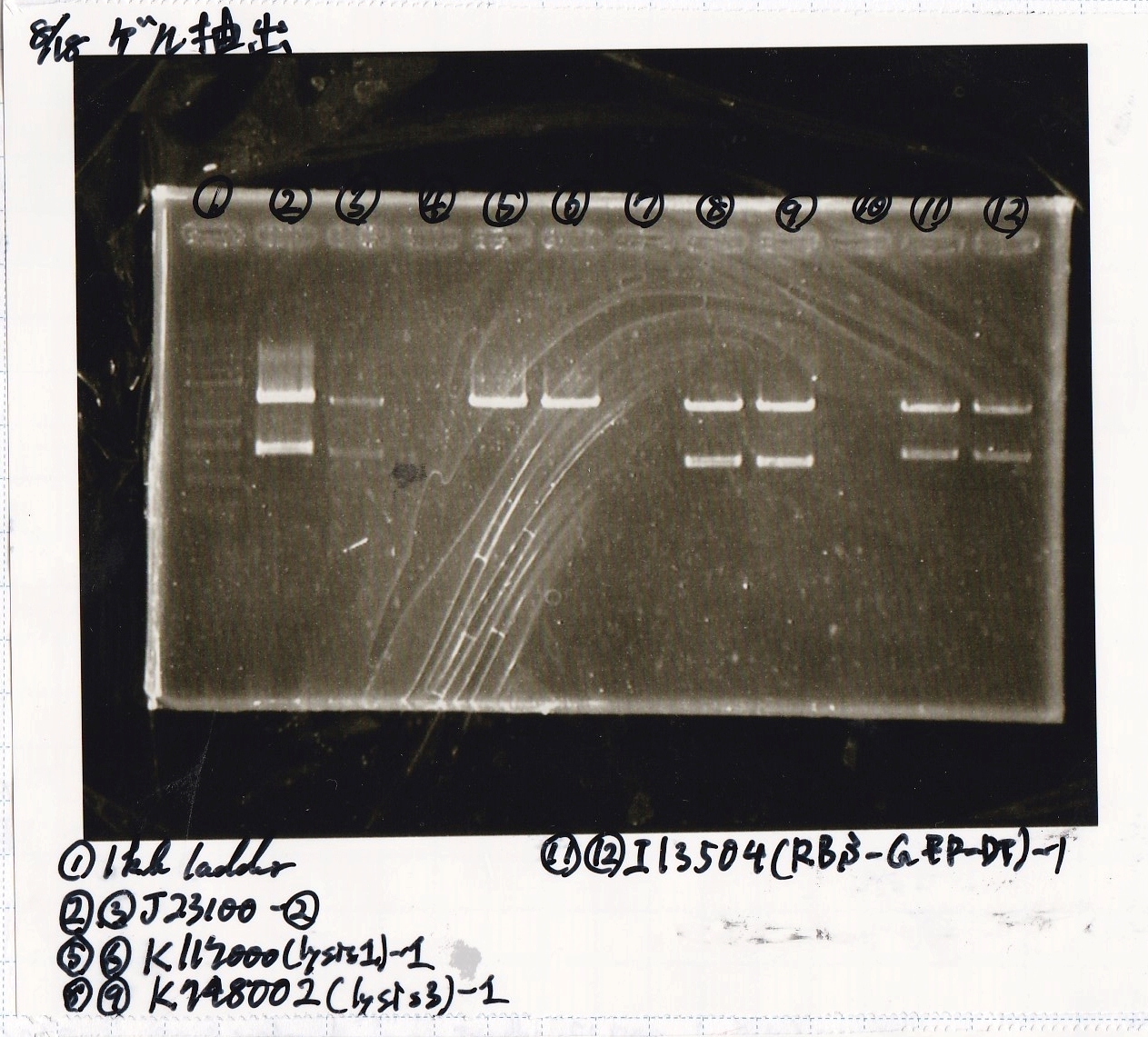
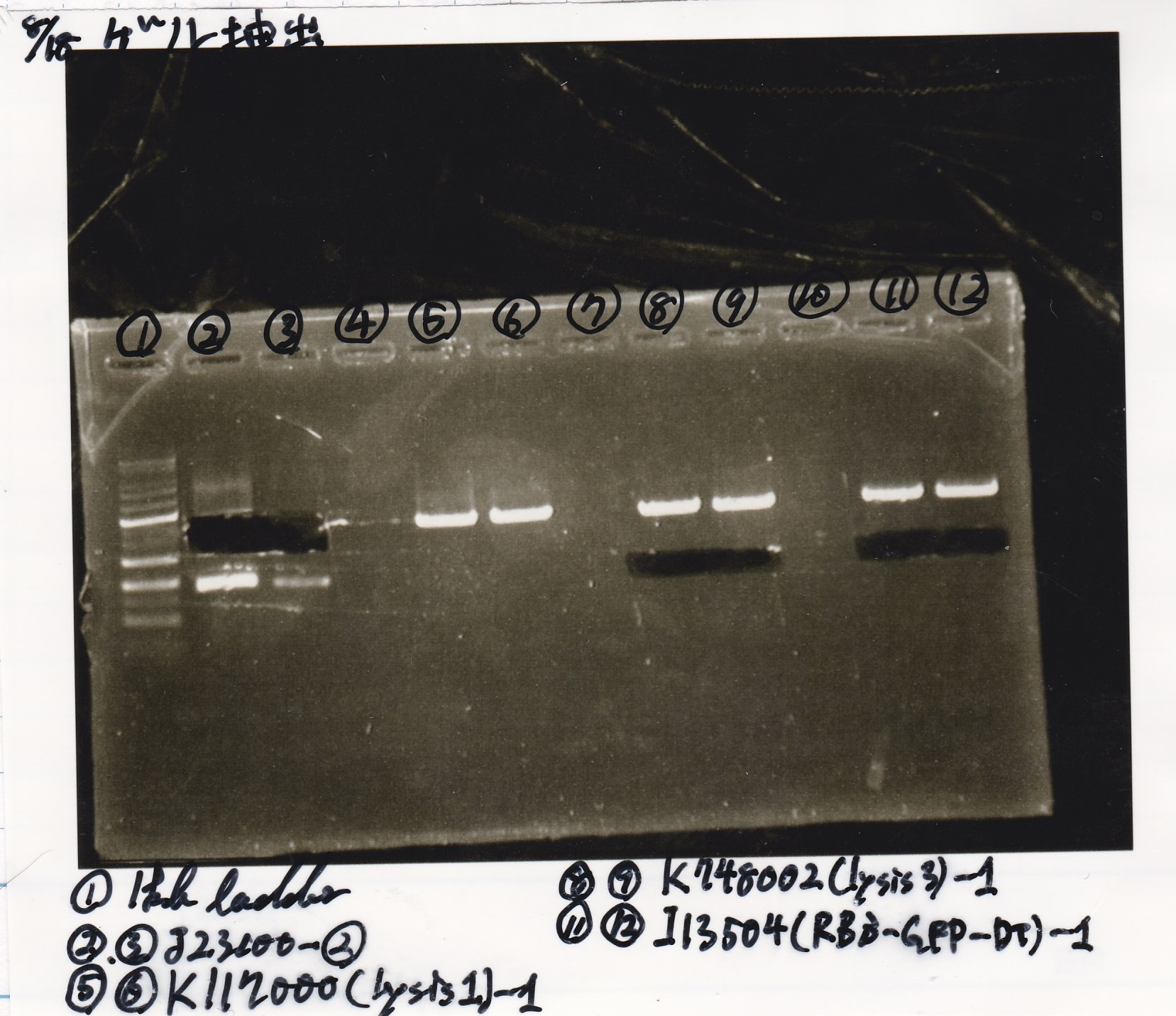
| Lane | DNA | Enzyme
|
| 13 | 1kb ladder | --
|
| 14 | 8/17 I0462(RBS-luxR-DT) -1 | --
|
| 15 | 8/17 I0462(RBS-luxR-DT) -1 | --
|
| 16 | -- | --
|
| 17 | 8/17 I732020(RBS-lacZα-DT) -1 | --
|
| 18 | 8/17 I732020(RBS-lacZα-DT) -1 | --
|
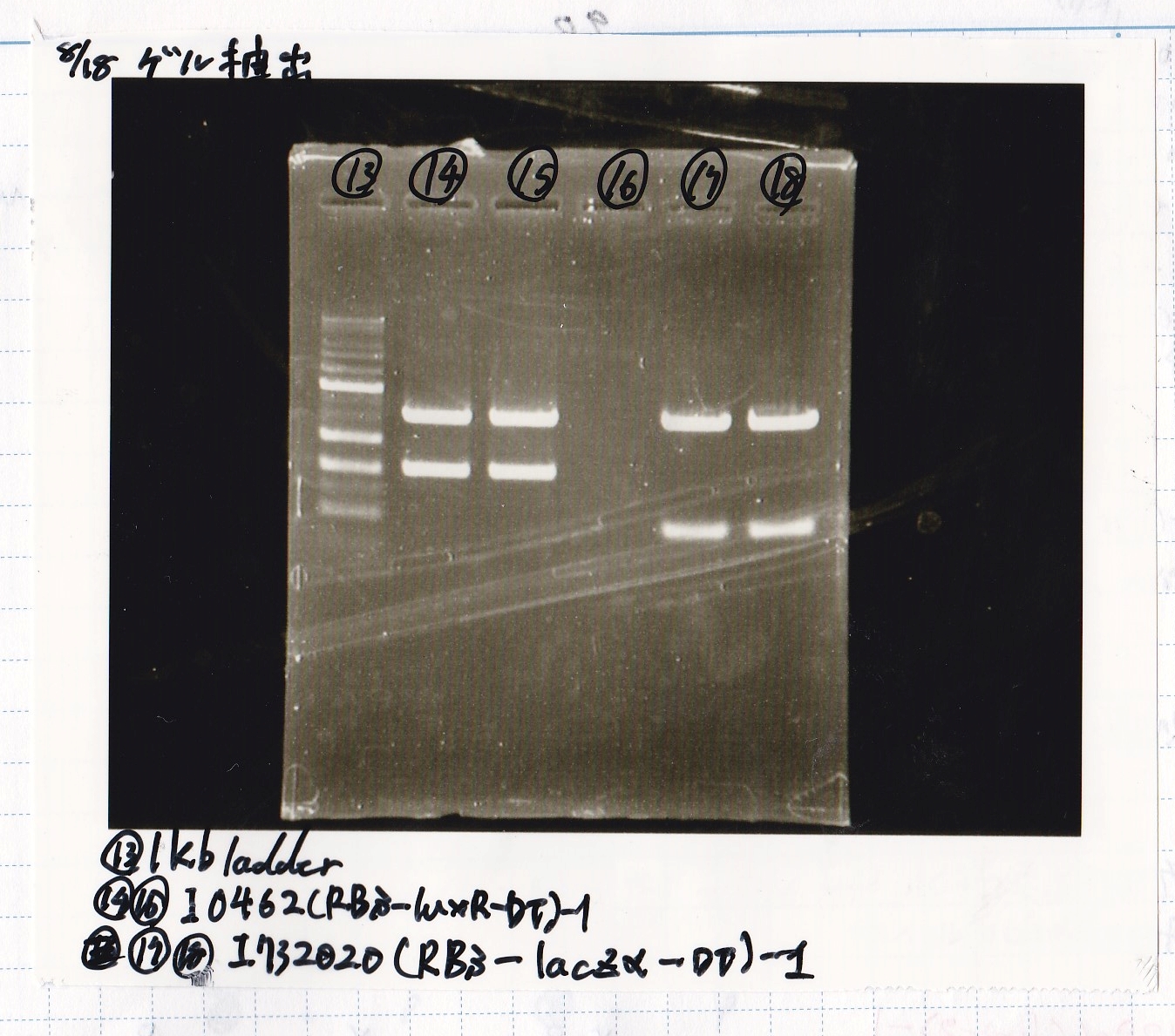
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/14 J231000 | 1.7 | 1.61 | 0.04
|
| 8/17 K748002(lysis3) -1 | 5.4 | 2.39 | 0.45
|
| 8/17 I13504(RBS-GFP-DT) -1 | 3.4 | 3.33 | 0.19
|
| 8/17 I0462(RBS-luxR-DT) -1 | 4.4 | 2.07 | 0.37
|
| 8/17 I732020(RBS-lacZα-DT) -1 | 2.9 | 2.17 | 0.29
|
Ligation
Ashida
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | J23100 | 9.0 | RBS-lacZα-DT | 8.6 | 3.5
|
| experiment | J23100 | 9.0 | RBS-GFP-DT | 7.3 | 3.5
|
| experiment | RBS | 1 | lysis3 | 4.6 | 2.8
|
| experiment | Plux | 1 | GFP | 7.3 | 3.5
|
| experiment | J23100 | 9.0 | -- | -- | 3.5
|
| experiment | RBS | 1.0 | -- | -- | 0.5
|
| experiment | Plux | 1.0 | -- | -- | 0.5
|
incubate 16°C 1hour
Restriction Enzyme Digestion
Murata and Okazaki
| | 8/17K112806(lysis2)-1 | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 2.0µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 3.0µL | 30 µL
|
| 1 cuts | 1.0&mic6/8ro;L | 0.2µL | -- | 1.0µL | 1.0µL | 6.8µL | 10 µL
|
| 1 cuts | 1.0µL | -- | 0.2µL | 1.0µL | 1.0µL | 6.8µL | 10 µL
|
Electrophoresis
Okazaki and Honda
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | lysis1 | XbaI | PstI
|
| 3 | lysis1 | XbaI | --
|
| 4 | lysis1 | -- | PstI
|
| 5 | lysis1 | -- | --
|
| 6 | lysis2 | XbaI | PstI
|
| 7 | lysis2 | XbaI | --
|
| 8 | lysis2 | -- | PstI
|
| 9 | lysis2 | -- | --
|
| 10 | 100bp ladder | -- | --
|
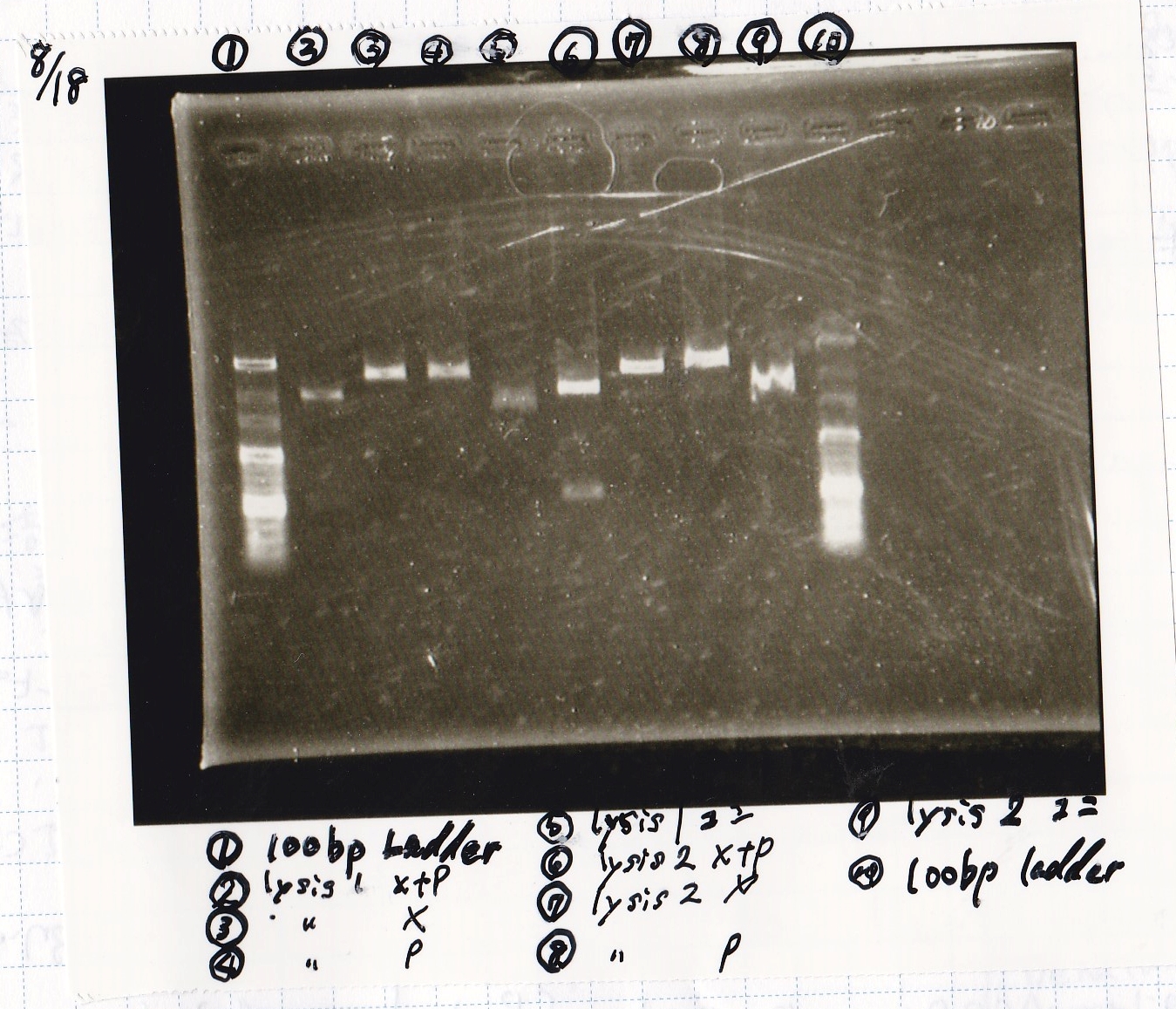
Colony PCR
No name
| Sample | base pair
|
| 8/8 LBKana | --
|
| 8/16 Pbad araC | --
|
| NC | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min | 30
|
Liquid Culture
No name
| Sample | medium
|
| 7/8 LB Kana | Plusgrow(+Kana)
|
| 8/16 Pbad araC | Plusgrow(+Kana)
|
LB Medium Plate
No name
| volume | 200ml
|
| Bacto Trypton | 2g
|
| Bacto yeasy extract | 1g
|
| NaCl | 1g
|
| Agar Powder | 2g
|
Transformation
No name
| Name | Sample | Competent Cells | Total | Plate
|
| 8/18 J23100+RBS-LacZα-DT | 2µL | 20µL | 22µL | Amp
|
| 8/18 J23100+RBS-GFP-DT | 2µL | 20µL | 22µL | Amp
|
| 8/18 J23100+RBS-luxR-DT | 2µL | 20µL | 22µL | Amp
|
| RBS+lysis3 | 2µL | 20µL | 22µL | Amp
|
| Plux+GFP | 2µL | 20µL | 22µL | CP
|
| J23100 | 2µL | 20µL | 22µL | Amp
|
| RBS | 2µL | 20µL | 22µL | Amp
|
| Plux | 2µL | 20µL | 22µL | CP
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | 8/16Pbad araC-1 | -- | --
|
| 3 | 8/16Pbad araC-2 | -- | --
|
| 4 | NC | -- | --
|
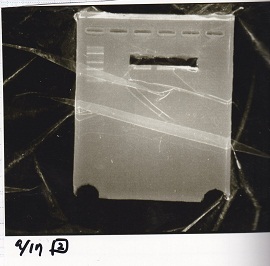
Aug 19
Colony PCR
Kojima
| Sample | base pair
|
| 8/18 J23100 control -(1) | 1142
|
| 8/18 J23100-RBS-GFP-DT -(1) | 1156
|
| 8/18 J23100-RBS-GFP-DT -(2) | 1156
|
| 8/18 J23100-RBS-luxR-DT -(1) | 1271
|
| 8/18 J23100-RBS-luxR-DT -(2) | 1271
|
| 8/18 J23100-RBS-lacZα-DT -(1) | 670
|
| 8/18 J23100-RBS-lacZα-DT -(2) | 670
|
| 8/18 RBS-lysis3 -(1) | 997
|
| 8/18 RBS-lysis3 -(2) | 997
|
| 8/18 P0440(RBS-tetR-DT) -(1) | 1154
|
| 8/18 P0440(RBS-tetR-DT) -(2) | 1154
|
| negative control | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min | 30cycles
|
| Lane | DNA
|
| 1 | 100bp ladder
|
| 2 | 8/18 NC J23100(1)
|
| 3 | 8/18 J23100-RBS-GFP-DT-(1)
|
| 4 | 8/18 J23100-RBS-GFP-DT-(2)
|
| 5 | 8/18 J23100-RBS-luxR-DT-(1)
|
| 6 | 8/18 J23100-RBS-luxR-DT-(2)
|
| 7 | 8/18 J23100-RBS-lacZα-DT-(1)
|
| 8 | 8/18 J23100-RBS-lacZα-DT-(2)
|
| 9 | 8/18 RBS-lysis3-(1)
|
| 10 | 8/18 RBS-lysis3-(2)
|
| 11 | 8/18 R0400(RBS-tetR-DT)-(1)
|
| 12 | 8/18 R0400(RBS-tetR-DT)-(2)
|
| 13 | NC
|
| 14 | 100bp ladder
|

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/19 lysis1 ① (XbaI & PstI) | 87.8 | 1.06 | 0.82
|
| Sample | base pair
|
| 8/18 RBS control -(1) | --
|
| 8/18 R0040(Ptet) -(1) | 686
|
| 8/18 R0040(Ptet) -(2) | 686
|
| negative control | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
Ashida
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 3 | 8/18 K117000(lysis2) -1 | XbaI & PstI
|
| 5 | 8/18 K117000(lysis2) -1 | XbaI & PstI
|
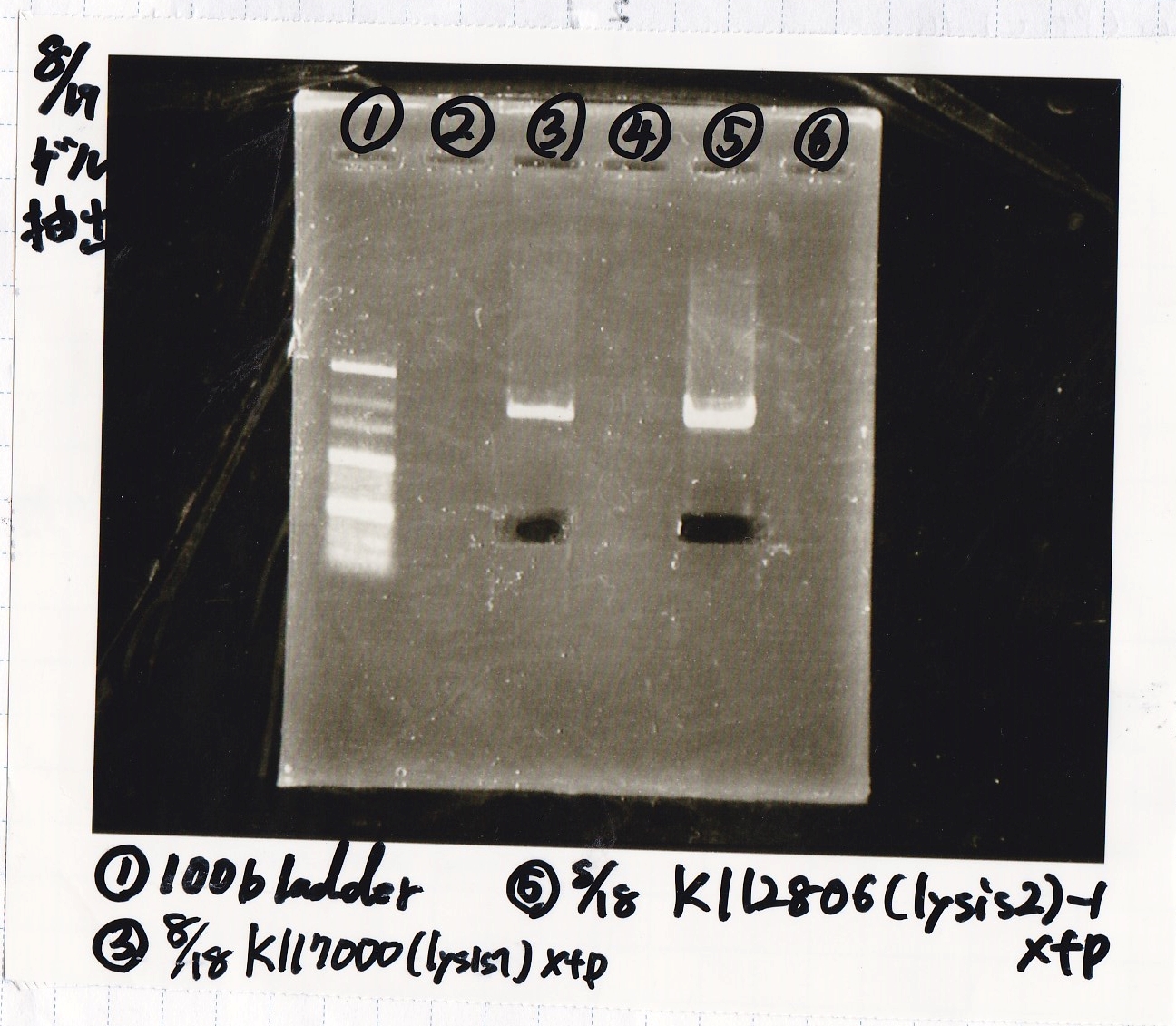
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/18 lysis2-1(XbaI & PstI) | 3.1 | 3.64 | 0.19
|
| 8/18 lysis2-1(XbaI & PstI) | 3.4 | 1.89 | 0.13
|
LB Medium Plate
Nakamoto
| volume | 200ml
|
| Bacto T2ypton | 2g
|
| Bacto yeast extract | 1g
|
| NaCl | 1g
|
| Agar Pouder | 2g
|
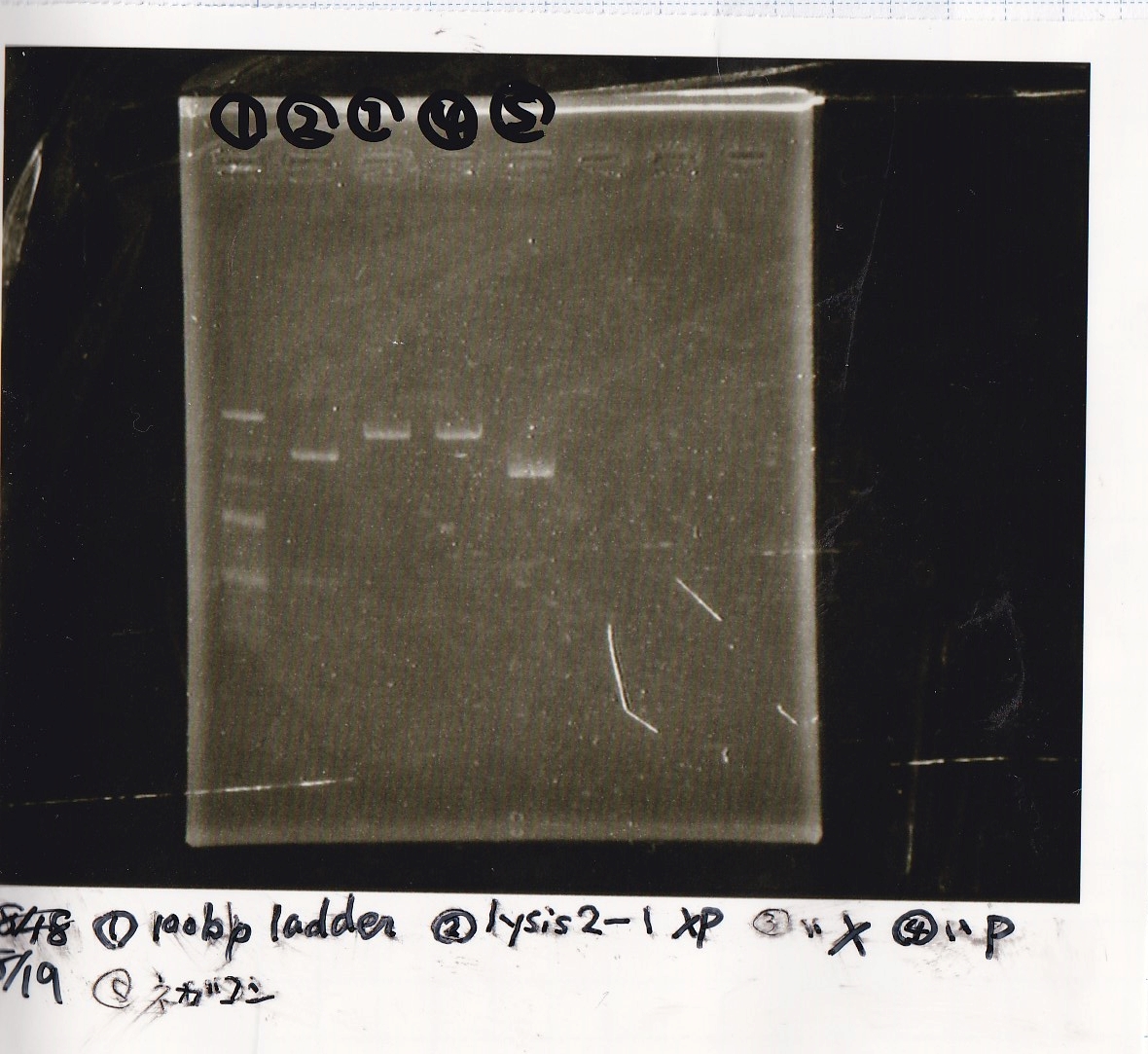
Restriction Enzyme Digestion
Tatsui
| | lysis2-1 | XbaI | PstI | 10xbuffer | BSA | MilliQ | total
|
| 2 cuts | 10µL | 1µL | 1µL | 3µL | 3µL | 12µL | 30µL
|
| 1 cut | 2µL | 0.2µL | -- | 1µL | 1µL | 5.8µL | 10µL
|
| 1 cut | 2µL | -- | 0.2µL | 1µL | 1µL | 5.8µL | 10µL
|
| NC | 2µL | -- | -- | 1µL | 1µ | 6µL | 10µL
|
Murakami and Okazaki
| | 8/19 lysis1-1 | XbaI | PstI | 10xbuffer | BSA | MilliQ | total
|
| 2 cuts | 2µL | 0.5µL | 0.8µL | 2µL | 2µL | 13µL | 20µL
|
| 1 cut | 0.5µL | 0.2µL | -- | 1µL | 1µL | 7.3µL | 10µL
|
| 1 cut | 0.5µL | -- | 0.2µL | 1µL | 1µL | 7.3µL | 10µL
|
| NC | 0.5µL | -- | -- | 1µL | 1µL | 7.5µL | 10µL
|
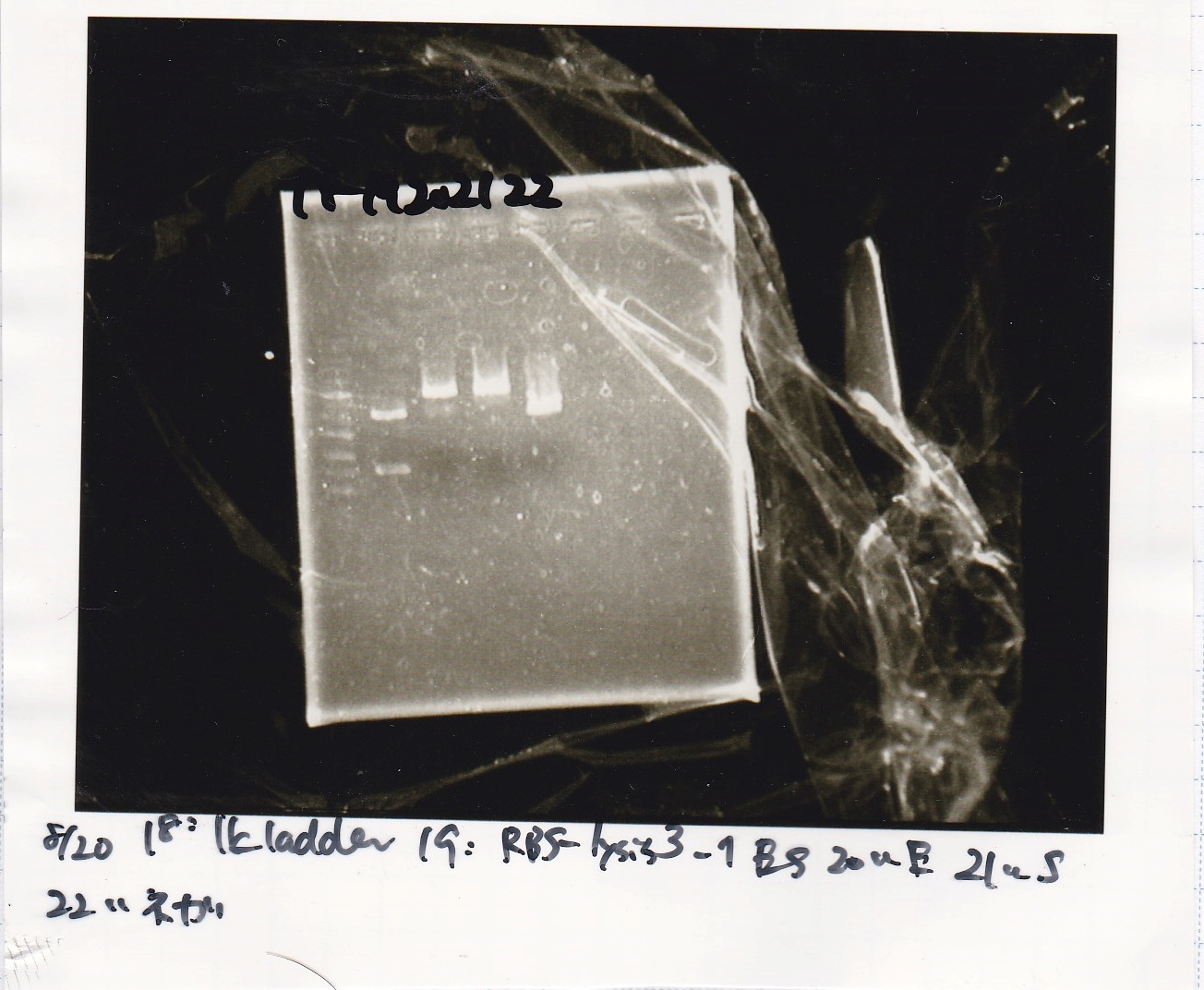
Ligation
No name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | RBS | 0.3 | lysis2 | 7.4 | 4
|
| NC | RBS | 0.3 | MilliQ | 7.4 | 4
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | 8/19 lysis1 | XbaI | PstI
|
| 3 | 8/19 lysis1 | XbaI | --
|
| 4 | 8/19 lysis1 | -- | PstI
|
| 5 | 8/19 lysis1 | -- | --
|
| 6 | 8/19 lysis1(Gel Extraction Product) | XbaI | PstI
|

Transformation
Murata and Kojima
| Name | Sample | Competent Cells | Plate
|
| 8/18 Plux+RBS-GFP-DT | 2µL | 20µL | CP
|
| 8/18 Plux(NC) | 2µL | 20µL | CP
|
| 8/19 RBS+lysis2 | 2µL | 20µL | Amp
|
| 8/15 Pbad-araC | 2µL | 20µL | Kan
|
No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | 8/19 lysis1 ① | XbaI & PstI
|
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/19 lysis1 ① (XbaI & PstI) | 87.8 | 1.06 | 0.82
|
Liquid Culture
Ashida
| Sample | medium
|
| 8/18 Ptet -1 | Plusgrow medium(+CP)
|
| 8/18 Ptet -2 | Plusgrow medium(+CP)
|
| 8/18 RBS-tetR-DT -1 | Plusgrow medium(+CP)
|
| 8/18 RBS-tetR-DT -2 | Plusgrow medium(+CP)
|
| 8/18 J23100-GFP -1 | Plusgrow medium(+Amp)
|
| 8/18 J23100-luxR -1 | Plusgrow medium(+Amp)
|
| 8/18 J23100-luxR -2 | Plusgrow medium(+Amp)
|
| 8/18 J23100-lacZα -1 | Plusgrow medium(+Amp)
|
| 8/18 RBS-lysis3 -1 | Plusgrow medium(+Amp)
|
| 8/18 RBS-lysis3 -2 | Plusgrow medium(+Amp)
|
Master Plate
Ashida
| Number | Use LB plate(+Amp)
|
| 1 | 8/18 J23100-GFP -1
|
| 2 | 8/18 J23100-luxR -1
|
| 3 | 8/18 J23100-luxR -2
|
| 4 | 8/18 J23100-lacZα -1
|
| 5 | 8/18 RBS-lysis3 -1
|
| 6 | 8/18 RBS-lysis3 -2
|
| Number | Use LB plate(+CP)
|
| 1 | 8/18 Ptet -1
|
| 1 | 8/18 Ptet -2
|
| 3 | 8/18 RBS-tetR-DT -1
|
| 4 | 8/18 RBS-tetR-DT -2
|
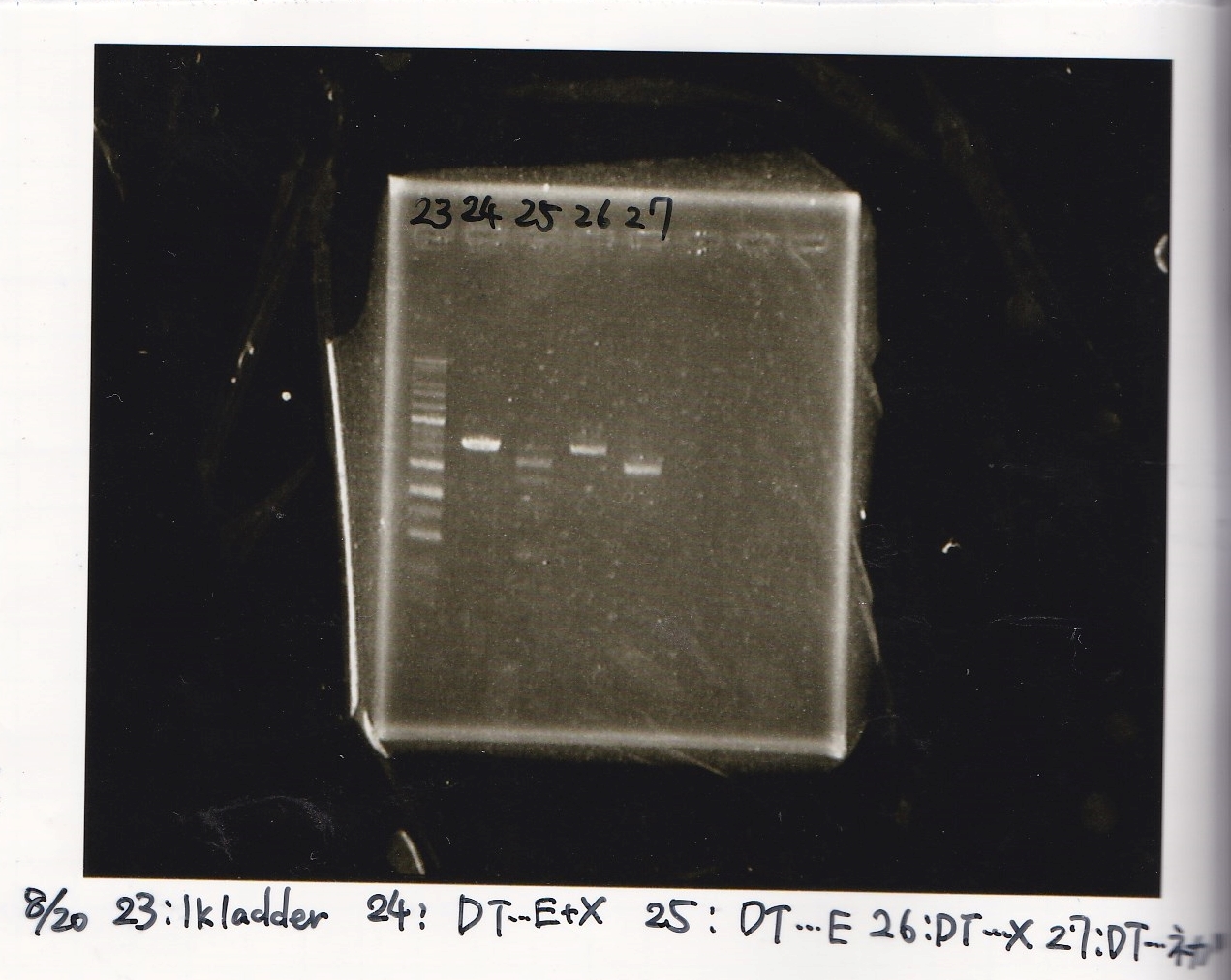
Aug 20
Miniprep
Kojima
| DNA | concentration[µg/µL] | 260/280 | 260/230
|
| 8/18 Ptet -1 | 220 | 1.69 | 1.87
|
| 8/18 Ptet -2 | 210 | 1.69 | 1.71
|
| 8/18 RBS-tetR-DT -1 | 236 | 1.63 | 1.69
|
| 8/18 RBS-tetR-DT -2 | 182 | 1.65 | 1.98
|
| 8/18 J23100-RBS-GFP-DT -1 | 334 | 1.71 | 2.12
|
| 8/18 J23100-RBS-luxR-DT -1 | 440 | 1.67 | 1.60
|
| 8/18 J23100-RBS-luxR-DT -2 | 344 | 1.68 | 1.83
|
| 8/18 J23100-RBS-lacZα-DT -1 | 280 | 1.70 | 1.81
|
| 8/18 RBS-lysis3 -1 | 282 | 1.67 | 1.76
|
| 8/18 RBS-lysis3 -2 | 212 | 1.30 | 1.35
|
LB Medium Plate
Okazaki
| volume | 200mL
|
| Bacto Trypton | 2g
|
| Bacto yeast extract | 1g
|
| Nacl | 1g
|
| Agar Powder | 2g
|
| Amp | 40µl
|
1. Measuring reagents and put they in the Erlenmeyer flask.
2. diluting in measuring cylinder to 200ml total.
3. autoclaving.
4. adding ampicirine 40ml.
Ristriction Enzyme Digestion
Kojima and Ashida
| | 8/20 Ptet -1(220µg/mL) | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total
|
| 2 cuts | 4.5 | 1 | 1 | 3 | 3 | 17.5 | 30
|
| 1 cut | 0.5 | 0.2 | 0 | 1 | 1 | 7.3 | 10
|
| 1 cut | 0.5 | 0 | 0.2 | 1 | 1 | 7.3 | 10
|
| NC | 0.5 | 0 | 0 | 1 | 1 | 7.5 | 10
|
| | 8/20 RBS-tetR-DT -2(182µg/mL) | EcoRI | SpeI | 10x buffer H | MilliQ | total
|
| 2 cuts | 5.5 | 1 | 1 | 3 | 19.5 | 30
|
| 1 cut | 0.6 | 0.2 | 0 | 1 | 8.2 | 10
|
| 1 cut | 0.6 | 0 | 0.2 | 1 | 8.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 8.4 | 10
|
| | 8/20 J23100-RBS-GFP-DT(334µg/ml) | EcoRI | SpeI | 10x buffer H | MilliQ | total
|
| 2 cuts | 3.0 | 1 | 1 | 3 | 22 | 30
|
| 1 cut | 0.3 | 0.2 | 0 | 1 | 8.5 | 10
|
| 1 cut | 0.3 | 0 | 0.2 | 1 | 8.5 | 10
|
| NC | 0.3 | 0 | 0 | 1 | 8.7 | 10
|
| | 8/20 J23100-RBS-luxR-DT -2(344µg/mL) | EcoRI | XbaI | BSA | 10x buffer M | MilliQ | total
|
| 2 cuts | 2.9 | 1 | 1 | 3 | 3 | 19.1 | 30
|
| 1 cut | 0.3 | 0.2 | 0 | 1 | 1 | 7.5 | 10
|
| 1 cut | 0.3 | 0 | 0.2 | 1 | 1 | 7.5 | 10
|
| NC | 0.3 | 0 | 0 | 1 | 1 | 7.7 | 10
|
| | 8/20 RBS-lysis3 -1(282µg/mL) | EcoRI | SpeI | 10x buffer H | MilliQ | total
|
| 2 cuts | 3.5 | 1 | 1 | 3 | 21.5 | 30
|
| 1 cut | 0.4 | 0.2 | 0 | 1 | 8.4 | 10
|
| 1 cut | 0.4 | 0 | 0.2 | 1 | 8.4 | 10
|
| NC | 0.4 | 0 | 0 | 1 | 8.6 | 10
|
| | 8/17 DT -1(188µg/mL) | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total
|
| 2 cuts | 5.3 | 1 | 1 | 3 | 3 | 16.7 | 30
|
| 1 cut | 0.3 | 0.2 | 0 | 1 | 1 | 7.5 | 10
|
| 1 cut | 0.3 | 0 | 0.2 | 1 | 1 | 7.5 | 10
|
| NC | 0.3 | 0 | 0 | 1 | 1 | 7.7 | 10
|
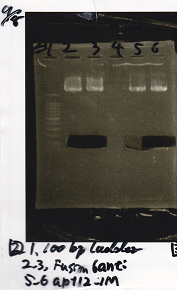
Transformation
Nakamoto
| Name | Sample | Competent Cells(XL10-gold) | Total | Plate
|
| tRNA-Spinach-tRNA | 1µL | 10µL | 11µL | Amp
|
| tetR-aptamer 12_PC(076) | 1µL | 10µL | 11µL | Amp
|
| tetR-aptamer 12_1R(113) | 1µL | 10µL | 11µL | Amp
|
| tetR-aptamer 12_1M(105) | 1µL | 10µL | 11µL | Amp
|
| pT181 attenuator | 1µL | 10µL | 11µL | Amp
|
| Fusin1 attenuator | 1µL | 10µL | 11µL | Amp
|
| Fusion3m2 attenuator | 1µL | 10µL | 11µL | Amp
|
| pT181 antisense | 1µL | 10µL | 11µL | Amp
|
| Fusion1 antisense | 1µL | 10µL | 11µL | Amp
|
| Fusion6 antisense | 1µL | 10µL | 11µL | Amp
|
incubate 37°C overnight 20:34~
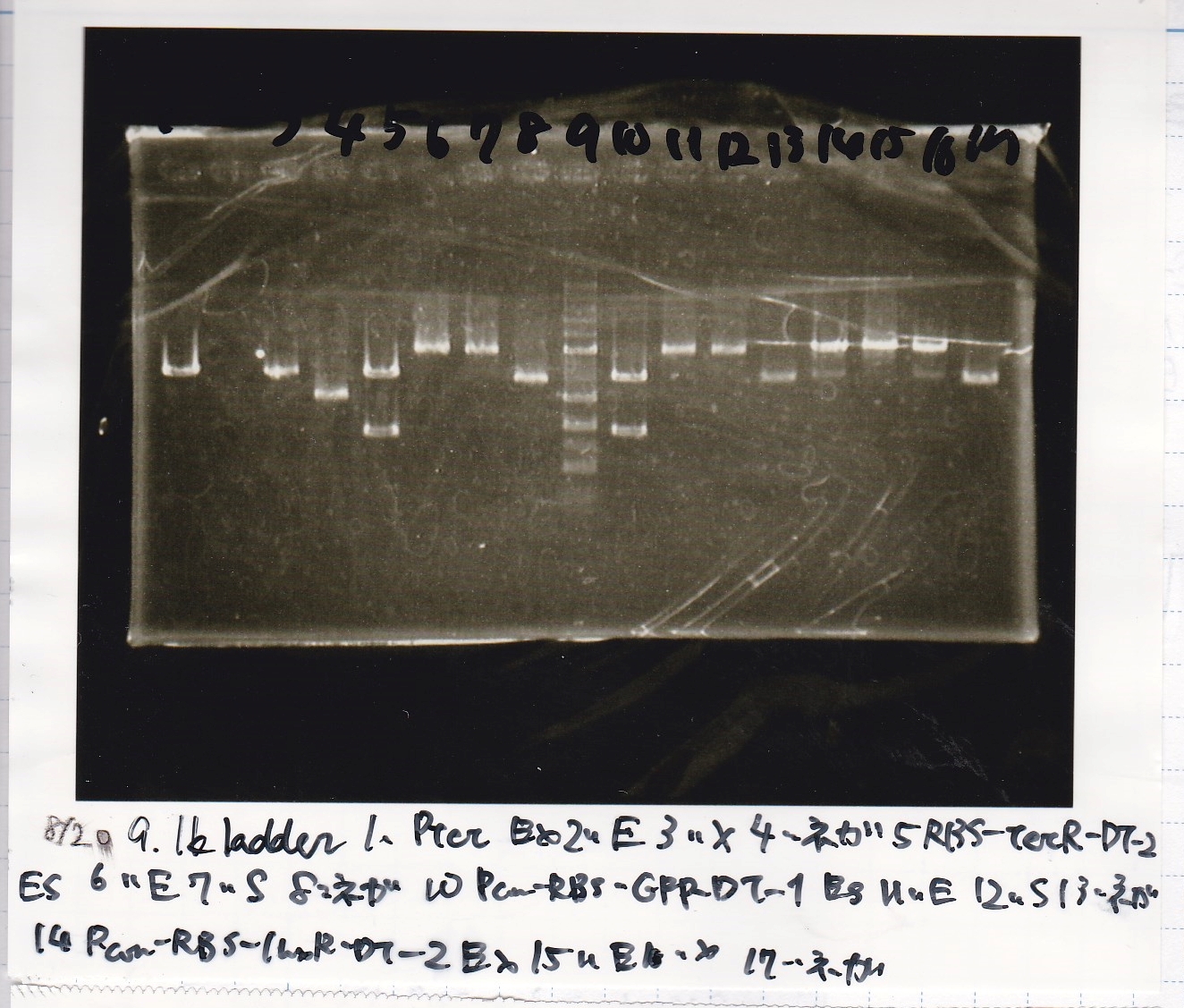
Electrophoresis
Kojima
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 8/20 Ptet -1 | EcoRI | XbaI
|
| 2 | 8/20 Ptet -1 | EcoRI | --
|
| 3 | 8/20 Ptet -1 | -- | XbaI
|
| 4 | 8/20 Ptet -1 | -- | --
|
| 5 | 8/20 RBS-tetR-DT -2 | EcoRI | SpeI
|
| 6 | 8/20 RBS-tetR-DT -2 | EcoRI | --
|
| 7 | 8/20 RBS-tetR-DT -2 | -- | SpeI
|
| 8 | 8/20 RBS-tetR-DT -2 | -- | --
|
| 9 | 1kbp ladder | -- | --
|
| 10 | 8/20 J23100-RBS-GFP-DT -1 | EcoRI | SpeI
|
| 11 | 8/20 J23100-RBS-GFP-DT -1 | EcoRI | --
|
| 12 | 8/20 J23100-RBS-GFP-DT -1 | -- | SpeI
|
| 13 | 8/20 J23100-RBS-GFP-DT -1 | -- | --
|
| 14 | 8/20 J23100-RBS-luxR-DT -1 | EcoRI | XbaI
|
| 15 | 8/20 J23100-RBS-luxR-DT -1 | EcoRI | --
|
| 16 | 8/20 J23100-RBS-luxR-DT -1 | -- | XbaI
|
| 17 | 8/20 J23100-RBS-luxR-DT -1 | -- | --
|
| 18 | 1kbp ladder | -- | --
|
| 19 | 8/20 RBS-lysis3 -1 | EcoRI | SpeI
|
| 20 | 8/20 RBS-lysis3 -1 | EcoRI | --
|
| 21 | 8/20 RBS-lysis3 -1 | -- | SpeI
|
| 22 | 8/20 RBS-lysis3 -1 | -- | --
|
| 23 | 1kbp ladder | -- | --
|
| 24 | 8/17 DT | EcoRI | XbaI
|
| 25 | 8/17 DT | EcoRI | --
|
| 26 | 8/17 DT | -- | XbaI
|
| 27 | 8/17 DT | -- | --
|
Aug 21
Plusgrow Medium
Tatsui
- 8/17 Plusgrow medium 50ml
- 7/22 5000x Ampicillin 10µl
- Measuring materials and putting them in a 50ml tube
Liquid culture
Tatsui
| Sample | medium
|
| 8/20 tRNA-spinach -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 tRNA-spinach -(2) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 TetR-aptamer 12_P(1076) -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 TetR-aptamer 12_P(1076) -(2) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 TetR-aptamer 12_1R(113) -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 TetR-aptamer 12_1R(113) -(2) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 TetR-aptamer 12_1M(105) -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 TetR-aptamer 12_1M(105) -(2) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 PT181 attenuator -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 PT181 attenuator -(2) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 Fusion1 attenuator -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 Fusion1 attenuator -(2) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 Fusion3m2 attenuator -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 Fusion3m2 attenuator -(2) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 PT181 antisense -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 PT181 antisense -(2) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 Fusion1 antisense -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 Fusion1 antisense -(2) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 Fusion6 antisense -(1) | 8/21 Plusgrow medium(+Amp)
|
| 8/20 Fusion6 antisense -(2) | 8/21 Plusgrow medium(+Amp)
|
Nakamoto and Tatsui
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 2 | 8/20 Ptet -(1) | EcoRI & XbaI
|
| 3
|
| 5 | 8/20 RBS-tetR-DT -(2) | EcoRI & SpeI
|
| 6
|
| 8 | 8/20 Pconst-RBS-GFP-DT -(1) | EcoRI & SpeI
|
| 9
|
| 11 | 8/20 RBS-lysis3 -(1) | EcoRI & SpeI
|
| 12
|
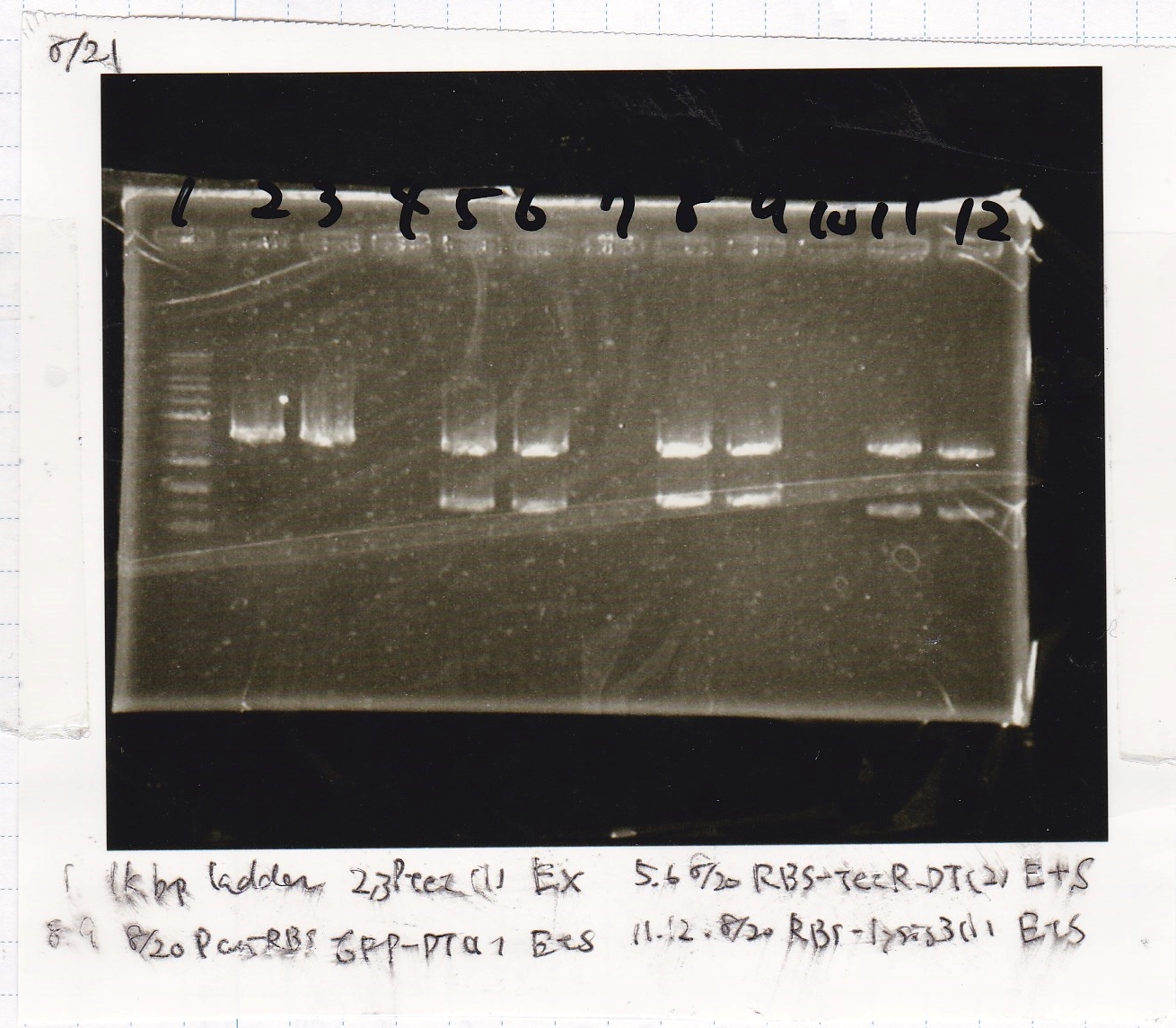

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Ptet (EcoRI & XbaI) | 21 | 1.55 | 0.78
|
| RBS-tetR-DT (EcoRI & SpeI) | 12 | 1.45 | 0.62
|
| Pconst-RBS-GFP-DT (EcoRI & SpeI) | 11 | 1.36 | 0.60
|
| RBS-lysis3 (EcoRI & SpeI) | 10 | 1.26 | 0.50
|
Ristriction Enzyme Digestion
Kojima and Nakamoto
| | 8/20 Ptet-(1) | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total
|
| 2 cuts | 4.5 | 1 | 1 | 3 | 3 | 17.5 | 30
|
| 1 cut | 0.5 | 0.2 | 0 | 1 | 1 | 7.3 | 10
|
| 1 cut | 0.5 | 0 | 0.2 | 1 | 1 | 7.3 | 10
|
| NC | 0.5 | 0 | 0 | 1 | 1 | 7.5 | 10
|
| | 8/20 Pconst-RBS-luxR-DT-(2) | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total
|
| 2 cuts | 2.9 | 1 | 1 | 3 | 3 | 19.1 | 30
|
| 1 cut | 0.3 | 0.2 | 0 | 1 | 1 | 7.5 | 10
|
| 1 cut | 0.3 | 0 | 0.2 | 1 | 1 | 7.5 | 10
|
| NC | 0.3 | 0 | 0 | 1 | 1 | 7.7 | 10
|
| | 8/17 DT | EcoRI | XbaI | 10x BSA | 10x buffer M | MilliQ | total
|
| 2 cuts | 5.3 | 1 | 1 | 3 | 3 | 16.7 | 30
|
| 1 cut | 0.5 | 0.2 | 0 | 1 | 1 | 7.3 | 10
|
| 1 cut | 0.5 | 0 | 0.2 | 1 | 1 | 7.3 | 10
|
| NC | 0.5 | 0 | 0 | 1 | 1 | 7.5 | 10
|
Electrophoresis
Kojima and Nakamoto
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kbp ladder | -- | --
|
| 2 | 8/20 Ptet -(1) | EcoRI | XbaI
|
| 3 | 8/20 Ptet -(1) | EcoRI | --
|
| 4 | 8/20 Ptet -(1) | -- | XbaI
|
| 5 | 8/20 Ptet -(1) | -- | --
|
| 6 | 8/20 J23100-RBS-luxR-DT -(2) | EcoRI | XbaI
|
| 7 | 8/20 J23100-RBS-luxR-DT -(2) | EcoRI | --
|
| 8 | 8/20 J23100-RBS-luxR-DT -(2) | -- | XbaI
|
| 9 | 8/20 J23100-RBS-luxR-DT -(2) | -- | --
|
| 10 | 8/20 DT | EcoRI | XbaI
|
| 11 | 8/20 DT | EcoRI | --
|
| 12 | 8/20 DT | -- | XbaI
|
| 13 | 8/20 DT | -- | --
|
| 14 | 1kbp ladder | -- | --
|
| 15 | 8/20 Ptet -(1) | EcoRI | XbaI
|
- The reason why lane 2 and 15 were same is that the well of lane 2 might be broken.

Kojima
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kbp ladder | -- | --
|
| 2 | 8/20 J23100-RBS-luxR-DT -(2) | EcoRI | XbaI
|
| 3 | 8/20 DT | EcoRI | XbaI
|

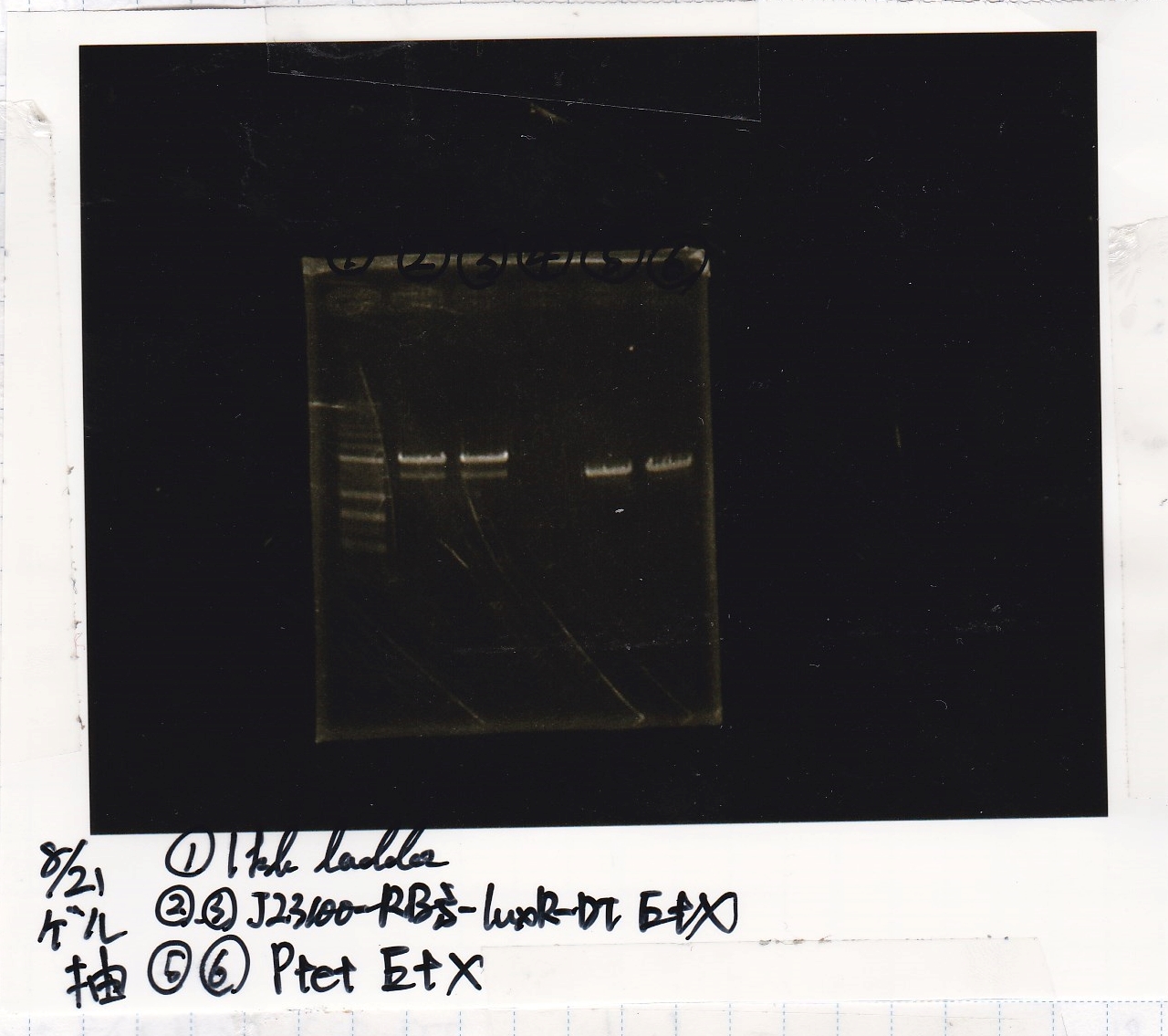
Honda
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 2 | 8/21 J23100-RBS-luxR-DT 25µL | EcoRI+XbaI
|
| 3 | 8/21 J23100-RBS-luxR-DT 25µL | EcoRI+XbaI
|
| 4 | -- | --
|
| 5 | 8/21 Ptet 25µL | EcoRI+XbaI
|
| 6 | 8/21 Ptet 25µL | EcoRI+XbaI
|

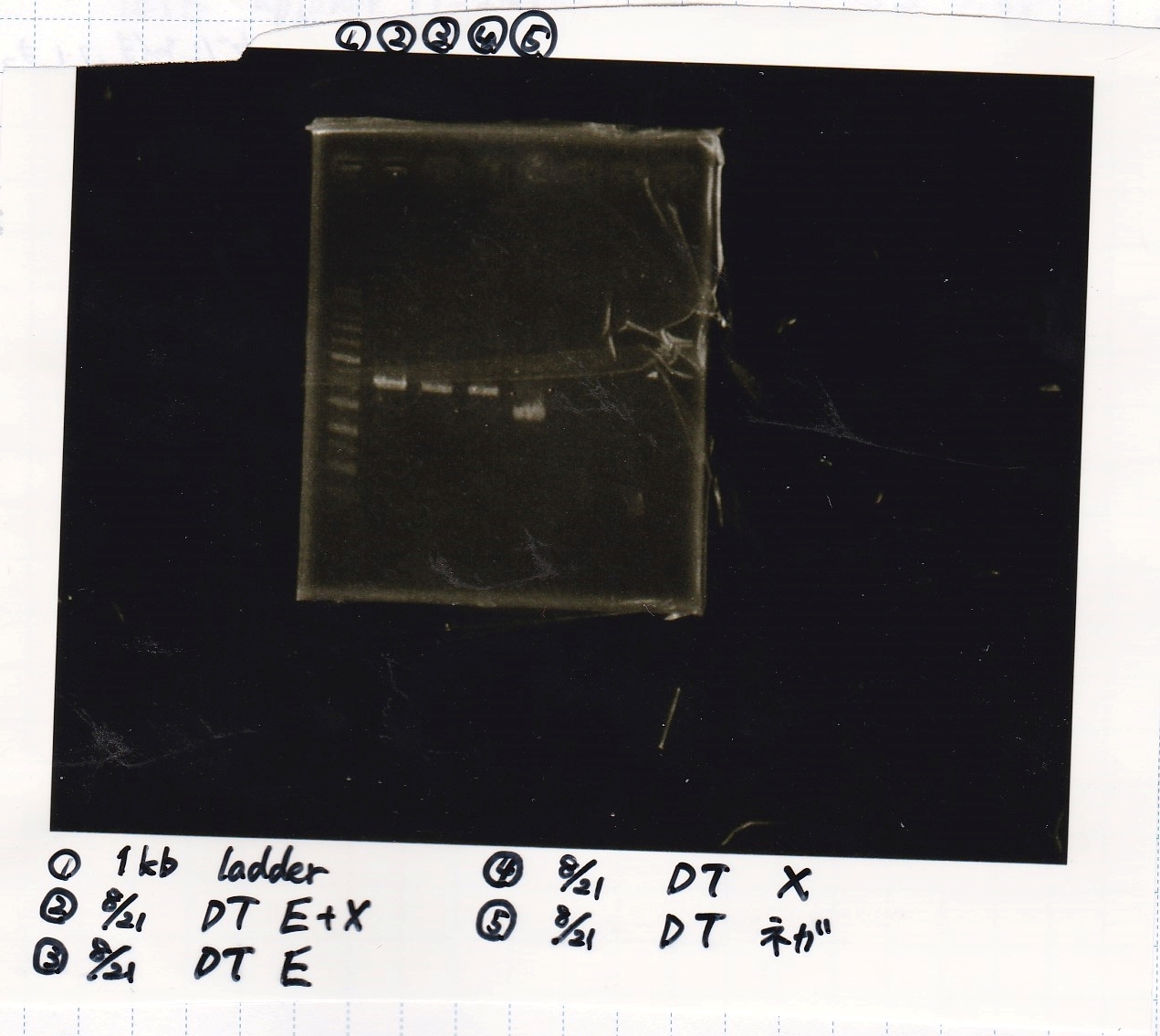
Electrophoresis
Honda
| Lane | Sample | EcoRI | XbaI
|
| 1 | 1kb ladder | -- | --
|
| 2 | 8/21 DT | + | +
|
| 3 | 8/21 DT | + | --
|
| 4 | 8/21 DT | -- | +
|
| 5 | 8/21 DT | -- | --
|
Ligation
No name
| state | Vector | Inserter | Ligation High ver.2 | total
|
| experiment | 8/21 Pcn-RBS-luxR-DT(+Amp) | 2.3 | 8/21 Pcn-RBS-GFP-DT | 1.7 | 2 | 6
|
| NC | 8/21 Pcn-RBS-luxR-DT(+Amp) | 2.3 | MilliQ | 1.7 | 2 | 6
|
| experiment | 8/18 Plux(+CP) | 1.1 | 8/19 RBS-GFP-DT | 5 | 3.1 | 9.2
|
| NC | 8/18 Plux(+CP) | 1.1 | MilliQ | 5 | 3.1 | 9.2
|
| experiment | 8/18 RBS(+Amp) | 2.2 | 8/19 lysis1 | 6.0 | 4.1 | 12.3
|
| experiment | 8/18 RBS(+Amp) | 2.2 | 8/19 lysis2 | 3.6 | 2.9 | 8.7
|
| NC | 8/18 RBS(+Amp) | 2.2 | MilliQ | 3.6 | 2.9 | 8.7
|
| experiment | 8/21 Ptet | 1.5 | 8/20 RBS-tetR-DT (2) | 2.1 | 1.8 | 5.4
|
| NC | 8/21 Ptet | 1.5 | MilliQ | 2.1 | 1.8 | 5.4
|
Transformation
Okazaki
| Name | Sample | Competent Cells | Total | Plate
|
| 8/21 Pcon-RBS-GFP-DT+Pcon-RBS-GFP-DT | 1µL | 10µL | 11µL | Amp
|
| 8/21 Pcon-RBS-GFP-DT NC | 1µL | 10µL | 11µL | Amp
|
| 8/18 RBS+8/19 lysis1 | 1µL | 10µL | 11µL | Amp
|
| 8/18 RBS+8/19 lysis2 | 1µL | 10µL | 11µL | Amp
|
| 8/18 RBS NC | 1µL | 10µL | 11µL | Amp
|
| 8/18 Plux+8/18 RBS-GFP-DT | 1µL | 10µL | 11µL | CP
|
| 8/18 Plux+8/18 RBS-GFP-DT | 1µL | 10µL | 11µL | CP
|
| 8/21 Ptet(pm)+8/20 RBS-tetR-DT(2) | 1µL | 10µL | 11µL | CP
|
| 8/21 Ptet(pm)+8/20 RBS-tetR-DT(2) NC | 1µL | 10µL | 11µL | CP
|
| 8/21 pSB1C3 | 1µL | 10µL | 11µL | CP
|
LB Medium Plate
Okazaki
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 2 | -- | --
|
| 3 | 8/21 DT | EcoRI & XbaI
|
| 4
|
Miniprep
Tatsui, Kojima, and Nakamoto
| DNA | concentration [µg/mL] | 260/280 | 260/230
|
| tRNA-spinach-(1) | 234 | 1.64 | 2.14
|
| tRNA-spinach-(2) | 440 | 1.60 | 1.91
|
| tetR-aptamer 12-p(1076)-(1) | 150 | 1.61 | 2.03
|
| tetR-aptamer 12-p(1076)-(2) | 374 | 1.45 | 1.66
|
| tetR-aptamer 12-1R(113)-(1) | 304 | 1.68 | 2.21
|
| tetR-aptamer 12-1R(113)-(2) | 382 | 1.47 | 1.66
|
| tetR-aptamer 12-1M(105)-(1) | 374 | 1.68 | 2.20
|
| tetR-aptamer 12-1M(105)-(2) | 286 | 1.31 | 1.43
|
| PT181 attenuator-(1) | 330 | 1.66 | 2.11
|
| PT181 attenuator-(2) | 270 | 1.68 | 2.18
|
| Fusion1 attenuator-(1) | 306 | 1.67 | 2.20
|
| Fusion1 attenuator-(2) | 282 | 1.60 | 1.94
|
| Fusion3m2 attenuator-(1) | 332 | 1.68 | 2.25
|
| Fusion3m2 attenuator-(2) | 326 | 1.67 | 2.10
|
| PT181 antisense-(1) | 178 | 1.64 | 1.68
|
| PT181 antisense-(2) | 116 | 1.63 | 1.95
|
| Fusion1 antisense-(1) | 300 | 1.67 | 2.21
|
| Fusion1 antisense-(2) | 164 | 1.42 | 1.46
|
| Fusion6 antisense-(1) | 336 | 1.65 | 2.18
|
| Fusion6 antisense-(2) | 192 | 1.65 | 2.01
|
Aug 22
Liquid culture
Nakamoto
| Sample | Medium
|
| 8/21 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT (1) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT (2) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 Pcon-RBS-GFP-DT control (1) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 Pcon-RBS-GFP-DT control (2) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 RBS-lysis1 (1) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 RBS-lysis1 (2) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 RBS control (1) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 RBS control (2) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 Ptet(pm)-RBS-tetR-DT (1) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 Ptet(pm)-RBS-tetR-DT (2) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 Ptet(pm) control (1) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 Ptet(pm) control (2) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 Plux-RBS-GFP-DT (1) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 pSB1C3(BBa_J04450) (1) | 8/21 Plusgrow medium (+Amp)
|
| 8/21 pSB1C3(BBa_J04450) (2) | 8/21 Plusgrow medium (+Amp)
|
incubate 37°C 10hour
Colony PCR
Kojima
| Sample | base pair
|
| 8/21 RBS-lysis1(1) | 400
|
| 8/21 RBS-lysis1(2) | 400
|
| 8/21 RBS control(1) | --
|
| 8/21 RBS control(2) | --
|
| negative control | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
| Sample | base pair
|
| 8/21 Pcon-GFP-DT-Pcon-RBS-luxR-DT (1) | 2143
|
| 8/21 Pcon-GFP-DT-Pcon-RBS-luxR-DT (2) | 2143
|
| 8/21 Pcon-RBS-luxR control(1) | --
|
| 8/21 Pcon-RBS-luxR control(2) | --
|
| negative control | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 2min | 30cycles
|
| Sample | base pair
|
| 8/21 Ptet-RBS-tetR-DT (1) | 1216
|
| 8/21 Ptet-RBS-tetR-DT (2) | 1216
|
| 8/21 Ptet control (1) | --
|
| 8/21 Ptet control (2) | --
|
| 8/21 Plux-RBS-GFP-DT (1) | 1227
|
| 8/21 pSB1C3(BBa_J04450) (1) | 1353
|
| 8/21 pSB1C3(BBa_J04450) (2) | 1353
|
| negative control | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min | 30cycles
|
Restriction Enzyme Digestion
Tatsui, Kojima, Honda and Ashida
| | Fusion1 antisense (1) (300 µg/mL) | EcoRI | SpeI | buffer H | MilliQ | total
|
| 2 cuts | 3.3 | 1 | 1 | 3 | 21.7 | 30
|
| 1 cut | 0.7 | 0.2 | 0 | 1 | 8.1 | 10
|
| 1 cut | 0.7 | 0 | 0.2 | 1 | 8.1 | 10
|
| NC | 0.7 | 0 | 0 | 1 | 8.3 | 10
|
| | Fusion1 antisense (1) (300 µg/mL) | XbaI | PstI | buffer M | BSA | MilliQ | total
|
| 2 cuts | 3.3 | 1 | 1 | 3 | 3 | 18.7 | 30
|
| 1 cut | 0.7 | 0.2 | 0 | 1 | 1 | 7.1 | 10
|
| 1 cut | 0.7 | 0 | 0.2 | 1 | 1 | 7.1 | 10
|
| NC | 0.7 | 0 | 0 | 1 | 1 | 7.3 | 10
|
| | Fusion6 antisense (1) (336 µg/mL) | EcoRI | SpeI | buffer H | MilliQ | total
|
| 2 cuts | 3 | 1 | 1 | 3 | 22 | 30
|
| 1 cut | 0.6 | 0.2 | 0 | 1 | 8.2 | 10
|
| 1 cut | 0.6 | 0 | 0.2 | 1 | 8.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 8.4 | 10
|
| | Fusion6 antisense (1) (336 µg/mL) | XbaI | PstI | buffer M | BSA | MilliQ | total
|
| 2 cuts | 3 | 1 | 1 | 3 | 3 | 19 | 30
|
| 1 cut | 0.6 | 0.2 | 0 | 1 | 1 | 7.2 | 10
|
| 1 cut | 0.6 | 0 | 0.2 | 1 | 1 | 7.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 1 | 7.4 | 10
|
| | pT181 antisense (1) (178 µg/mL) | EcoRI | SpeI | buffer H | MilliQ | total
|
| 2 cuts | 5.6 | 1 | 1 | 3 | 19.4 | 30
|
| 1 cut | 1.1 | 0.2 | 0 | 1 | 7.7 | 10
|
| 1 cut | 1.1 | 0 | 0.2 | 1 | 7.7 | 10
|
| NC | 1.1 | 0 | 0 | 1 | 7.9 | 10
|
| | pT181 antisense (1) (178 µg/mL) | XbaI | PstI | buffer M | BSA | MilliQ | total
|
| 2 cuts | 5.6 | 1 | 1 | 3 | 3 | 16.4 | 30
|
| 1 cut | 1.1 | 0.2 | 0 | 1 | 1 | 6.7 | 10
|
| 1 cut | 1.1 | 0 | 0.2 | 1 | 1 | 6.7 | 10
|
| NC | 1.1 | 0 | 0 | 1 | 1 | 6.9 | 10
|
| | pT181 attenuator (1) | EcoRI | SpeI | buffer H | MilliQ | total
|
| 2 cuts | 3 | 1 | 1 | 3 | 22 | 30
|
| 1 cut | 0.6 | 0.2 | 0 | 1 | 8.2 | 10
|
| 1 cut | 0.6 | 0 | 0.2 | 1 | 8.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 8.4 | 10
|
| | pT181 attenuator (1) | XbaI | PstI | buffer M | BSA | MilliQ | total
|
| 2 cuts | 3 | 1 | 1 | 3 | 3 | 19 | 30
|
| 1 cut | 0.6 | 0.2 | 0 | 1 | 1 | 7.2 | 10
|
| 1 cut | 0.6 | 0 | 0.2 | 1 | 1 | 7.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 1 | 7.4 | 10
|
| | Fusion1 attenuator (1) | EcoRI | SpeI | buffer H | MilliQ | total
|
| 2 cuts | 3.3 | 1 | 1 | 3 | 21.7 | 30
|
| 1 cut | 0.7 | 0.2 | 0 | 1 | 8.1 | 10
|
| 1 cut | 0.7 | 0 | 0.2 | 1 | 8.1 | 10
|
| NC | 0.7 | 0 | 0 | 1 | 8.3 | 10
|
| | Fusion1 attenuator (1) | XbaI | PstI | buffer M | BSA | MilliQ | total
|
| 2 cuts | 3.3 | 1 | 1 | 3 | 3 | 18.7 | 30
|
| 1 cut | 0.7 | 0.2 | 0 | 1 | 1 | 7.1 | 10
|
| 1 cut | 0.7 | 0 | 0.2 | 1 | 1 | 7.1 | 10
|
| NC | 0.7 | 0 | 0 | 1 | 1 | 7.3 | 10
|
| | Fusion3m2 attenuator (1) | EcoRI | SpeI | buffer H | MilliQ | total
|
| 2 cuts | 3 | 1 | 1 | 3 | 22 | 30
|
| 1 cut | 0.6 | 0.2 | 0 | 1 | 8.2 | 10
|
| 1 cut | 0.6 | 0 | 0.2 | 1 | 8.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 8.4 | 10
|
| | Fusion3m2 attenuator (1) | XbaI | PstI | buffer M | BSA | MilliQ | total
|
| 2 cuts | 3 | 1 | 1 | 3 | 3 | 19 | 30
|
| 1 cut | 0.6 | 0.2 | 0 | 1 | 1 | 7.2 | 10
|
| 1 cut | 0.6 | 0 | 0.2 | 1 | 1 | 7.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 1 | 7.4 | 10
|
| | Spinach (2) | EcoRI | SpeI | buffer B | BSA | MilliQ | total
|
| 2 cuts | 2.3 | 0.5 | 0.5 | 3 | 0.3 | 23.4 | 30
|
| 1 cut | 0.5 | 0.1 | 0 | 1 | 0.1 | 8.3 | 10
|
| 1 cut | 0.5 | 0 | 0.1 | 1 | 0.1 | 8.3 | 10
|
| NC | 0.5 | 0 | 0 | 1 | 0.1 | 8.4 | 10
|
| | tetR aptamer 12_P (2) | EcoRI | SpeI | buffer B | BSA | MilliQ | total
|
| 2 cuts | 2.7 | 0.5 | 0.5 | 3 | 0.3 | 23 | 30
|
| 1 cut | 0.5 | 0.1 | 0 | 1 | 0.1 | 8.3 | 10
|
| 1 cut | 0.5 | 0 | 0.1 | 1 | 0.1 | 8.3 | 10
|
| NC | 0.5 | 0 | 0 | 1 | 0.1 | 8.4 | 10
|
| | tetR aptamer 12_1R (2) | EcoRI | SpeI | buffer B | BSA | MilliQ | total
|
| 2 cuts | 2.6 | 0.5 | 0.5 | 3 | 0.3 | 23.1 | 30
|
| 1 cut | 0.5 | 0.1 | 0 | 1 | 0.1 | 8.3 | 10
|
| 1 cut | 0.5 | 0 | 0.1 | 1 | 0.1 | 8.3 | 10
|
| NC | 0.5 | 0 | 0 | 1 | 0.1 | 8.4 | 10
|
| | tetR aptamer 12_1M (1) | EcoRI | SpeI | buffer B | BSA | MilliQ | total
|
| 2 cuts | 2.7 | 0.5 | 0.5 | 3 | 0.3 | 23 | 30
|
| 1 cut | 0.5 | 0.1 | 0 | 1 | 0.1 | 8.3 | 10
|
| 1 cut | 0.5 | 0 | 0.1 | 1 | 0.1 | 8.3 | 10
|
| NC | 0.5 | 0 | 0 | 1 | 0.1 | 8.4 | 10
|
| | J23100 | SpeI | PstI | buffer B | BSA | MilliQ | total
|
| 2 cuts | 7 | 0.5 | 0.5 | 3 | 0.3 | 18.7 | 30
|
| 1 cut | 1.4 | 0.1 | 0 | 1 | 0.1 | 7.4 | 10
|
| 1 cut | 1.4 | 0 | 0.1 | 1 | 0.1 | 7.4 | 10
|
| NC | 1.4 | 0 | 0 | 1 | 0.1 | 7.5 | 10
|
| | Plac (2) | SpeI | PstI | buffer B | BSA | MilliQ | total
|
| 2 cuts | 11.2 | 0.5 | 0.5 | 3 | 0.3 | 14.5 | 30
|
| 1 cut | 2.2 | 0.1 | 0 | 1 | 0.1 | 6.6 | 10
|
| 1 cut | 2.2 | 0 | 0.1 | 1 | 0.1 | 6.6 | 10
|
| NC | 2.2 | 0 | 0 | 1 | 0.1 | 6.7 | 10
|
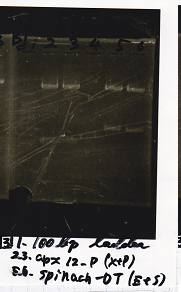
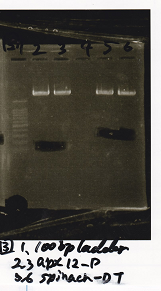
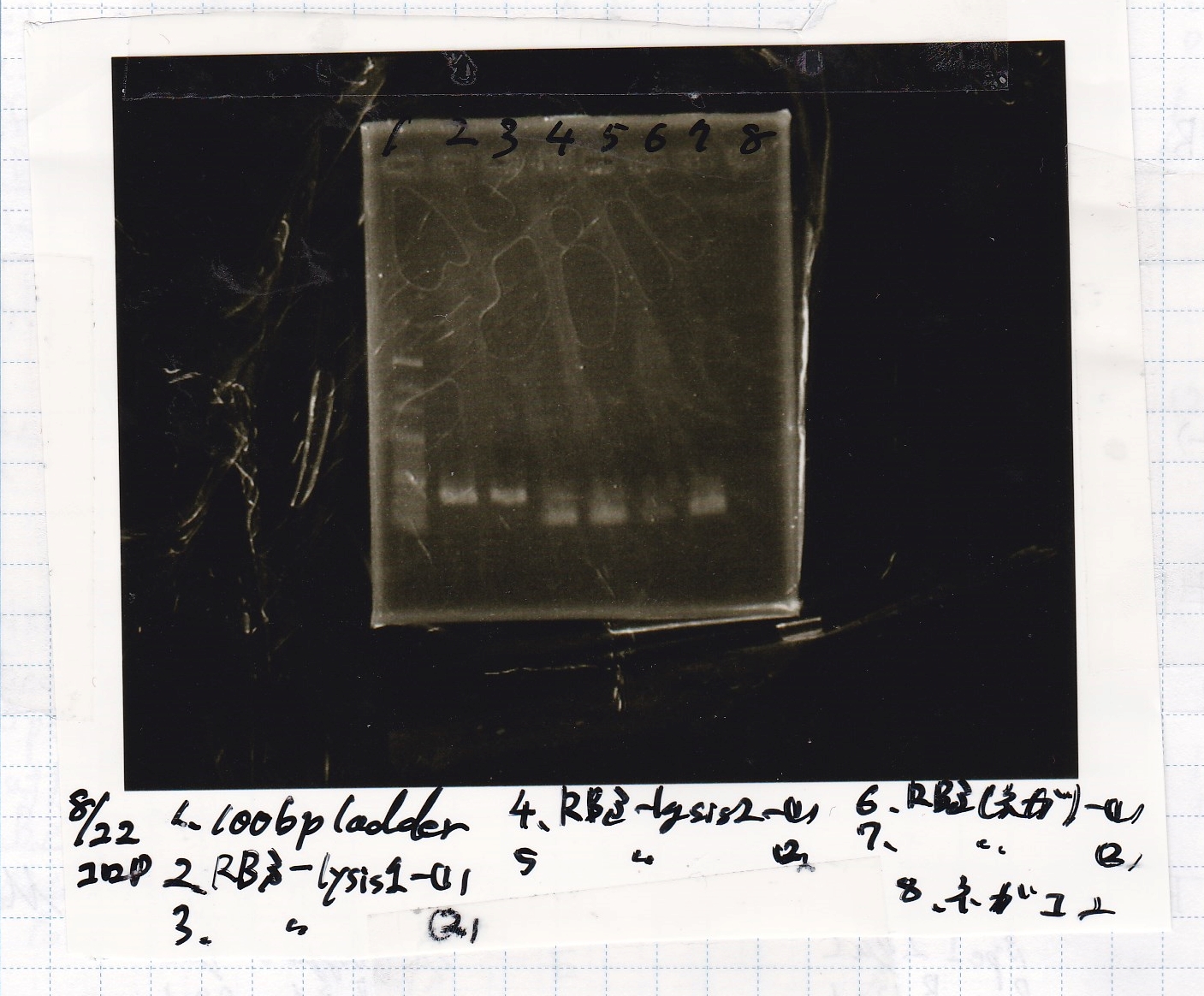
Electrophoresis
No name
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | RBS-lysis1 -(1)
|
| 3 | RBS-lysis1 -(2)
|
| 4 | RBS-lysis2 -(1)
|
| 5 | RBS-lysis2 -(2)
|
| 6 | RBS(NC) -(1)
|
| 7 | RBS(NC) -(2)
|
| 8 | NC
|
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | Ptet-RBS-tetR-DT -(1)
|
| 3 | Ptet-RBS-tetR-DT -(2)
|
| 4 | Ptet(NC) -(1)
|
| 5 | Ptet(NC) -(2)
|
| 6 | Plux-RBS-GFP-DT -(1)
|
| 7 | pSB1C3 -(1)
|
| 8 | pSB1C3 -(2)
|
| 9 | NC
|
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT -(1)
|
| 3 | Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT -(2)
|
| 4 | Pcon-RBS-luxR -(1)
|
| 5 | Pcon-RBS-luxR -(2)
|
| 6 | NC
|
| 7 | 1kb ladder
|

Miniprep
Ashida
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 8/22 RBS-lysis (1) | 128 | 1.77 | 2.29
|
| 8/22 RBS-lysis (2) | 96 | 1.80 | 2.07
|
| 8/22 pSB1C3 (1) | 108 | 1.55 | 1.45
|
| 8/22 pSB1C3 (2) | 130 | 1.69 | 1.76
|
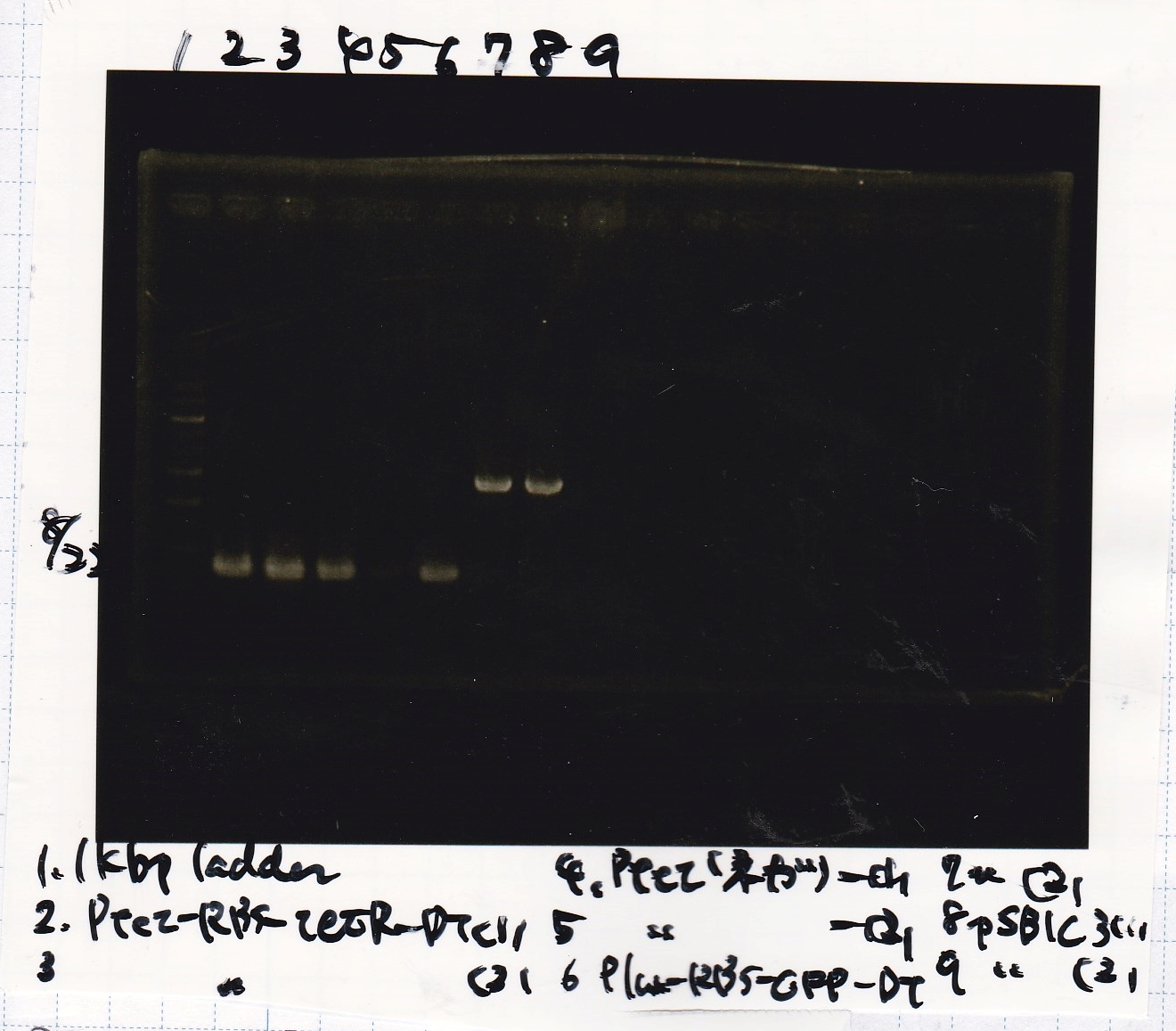
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp | -- | --
|
| 2 | 8/22 pT181 attenuator (1) | EcoRI | SpeI
|
| 3 | 8/22 pT181 attenuator (1) | EcoRI | --
|
| 4 | 8/22 pT181 attenuator (1) | -- | SpeI
|
| 5 | 8/22 pT181 attenuator (1) | -- | --
|
| 6 | 8/22 pT181 attenuator (1) | XbaI | PstI
|
| 7 | 8/22 pT181 attenuator (1) | XbaI | --
|
| 8 | 8/22 pT181 attenuator (1) | -- | PstI
|
| 9 | 8/22 pT181 attenuator (1) | -- | --
|
| 10 | 8/22 Fusion6 antisense (1) | EcoRI | SpeI
|
| 11 | 8/22 Fusion6 antisense (1) | EcoRI | --
|
| 12 | 8/22 Fusion6 antisense (1) | -- | SpeI
|
| 13 | 8/22 Fusion6 antisense (1) | -- | --
|
| 14 | 100bp | -- | --
|
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp | -- | --
|
| 2 | 8/22 Fusion6 antisense (1) | XbaI | PstI
|
| 3 | 8/22 Fusion6 antisense (1) | XbaI | --
|
| 4 | 8/22 Fusion6 antisense (1) | -- | PstI
|
| 5 | 8/22 Fusion6 antisense (1) | -- | --
|
| 6 | 8/22 Fusion1 antisense (1) | EcoRI | SpeI
|
| 7 | 8/22 Fusion1 antisense (1) | EcoRI | --
|
| 8 | 8/22 Fusion1 antisense (1) | -- | SpeI
|
| 9 | 8/22 Fusion1 antisense (1) | -- | --
|
| 10 | 8/22 Fusion1 antisense (1) | XbaI | PstI
|
| 11 | 8/22 Fusion1 antisense (1) | XbaI | --
|
| 12 | 8/22 Fusion1 antisense (1) | -- | PstI
|
| 13 | 8/22 Fusion1 antisense (1) | -- | --
|
| 14 | 100bp | -- | --
|
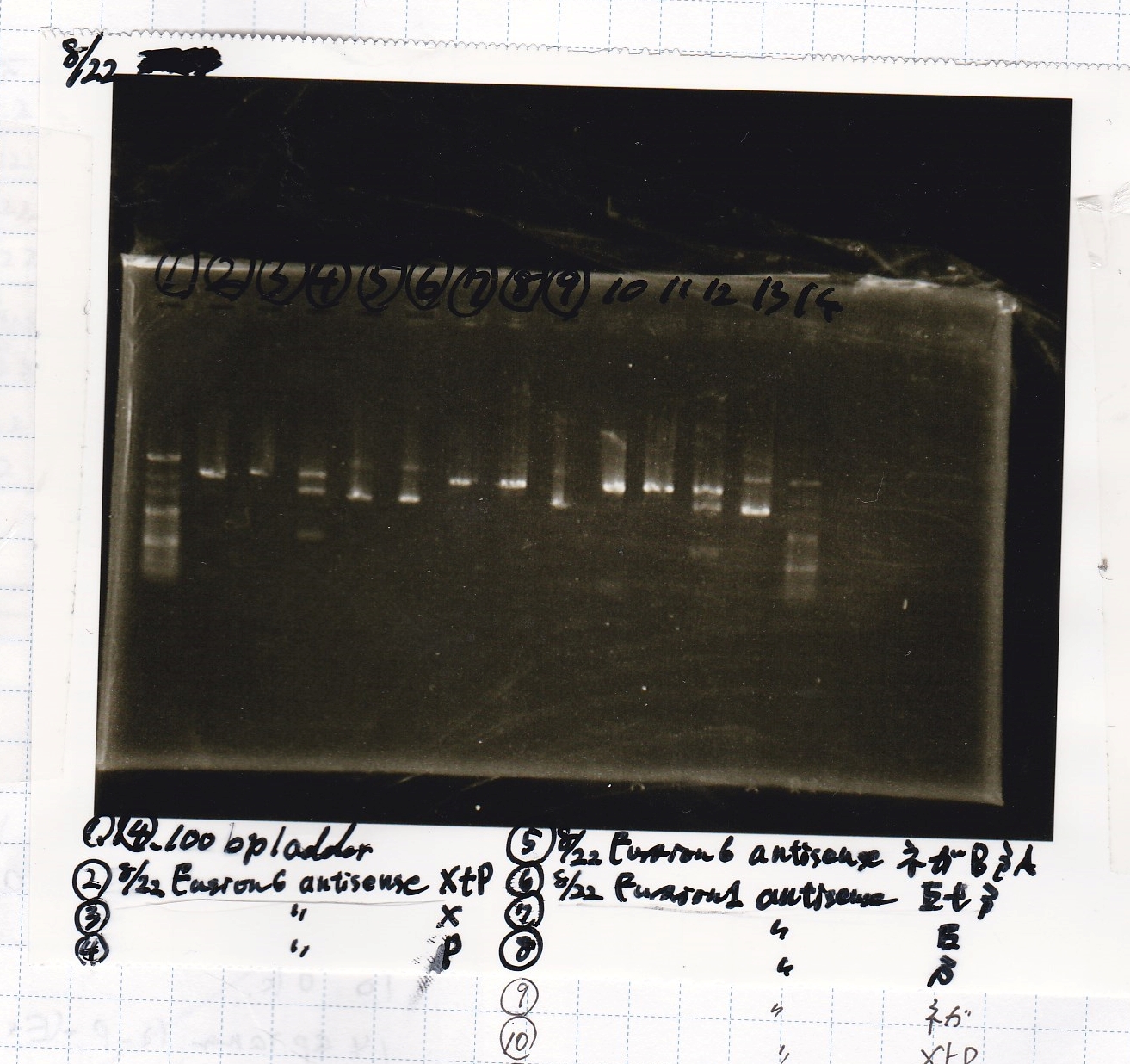
Aug 23
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | pT181 antisense | EcoRI | SpeI
|
| 2 | pT181 antisense | EcoRI | --
|
| 3 | pT181 antisense | -- | SpeI
|
| 4 | pT181 antisense | -- | --
|
| 5 | pT181 antisense | XbaI | PstI
|
| 6 | pT181 antisense | XbaI | --
|
| 7 | pT181 antisense | -- | PstI
|
| 8 | pT181 antisense | -- | --
|
| 9 | 100bp ladder | -- | --
|
| 10 | Fusion1 attenuator | EcoRI | SpeI
|
| 11 | Fusion1 attenuator | EcoRI | --
|
| 12 | Fusion1 attenuator | -- | SpeI
|
| 13 | Fusion1 attenuator | -- | --
|
| 14 | Fusion1 attenuator | XbaI | PstI
|
| 15 | Fusion1 attenuator | XbaI | --
|
| 16 | Fusion1 attenuator | -- | PstI
|
| 17 | Fusion1 attenuator | -- | --
|

| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | Fusion3m2 attenuator | EcoRI | SpeI
|
| 2 | Fusion3m2 attenuator | EcoRI | --
|
| 3 | Fusion3m2 attenuator | -- | SpeI
|
| 4 | Fusion3m2 attenuator | -- | --
|
| 5 | Fusion3m2 attenuator | XbaI | PstI
|
| 6 | Fusion3m2 attenuator | XbaI | --
|
| 7 | Fusion3m2 attenuator | -- | PstI
|
| 8 | Fusion3m2 attenuator | -- | --
|
| 9 | 100bp ladder | -- | --
|
| 10 | tetR aptamer 12_1M | EcoRI | SpeI
|
| 11 | tetR aptamer 12_1M | EcoRI | --
|
| 12 | tetR aptamer 12_1M | -- | SpeI
|
| 13 | tetR aptamer 12_1M | -- | --
|
| 14 | tetR aptamer 12_P | EcoRI | SpeI
|
| 15 | tetR aptamer 12_P | EcoRI | --
|
| 16 | tetR aptamer 12_P | -- | SpeI
|
| 17 | tetR aptamer 12_P | -- | --
|
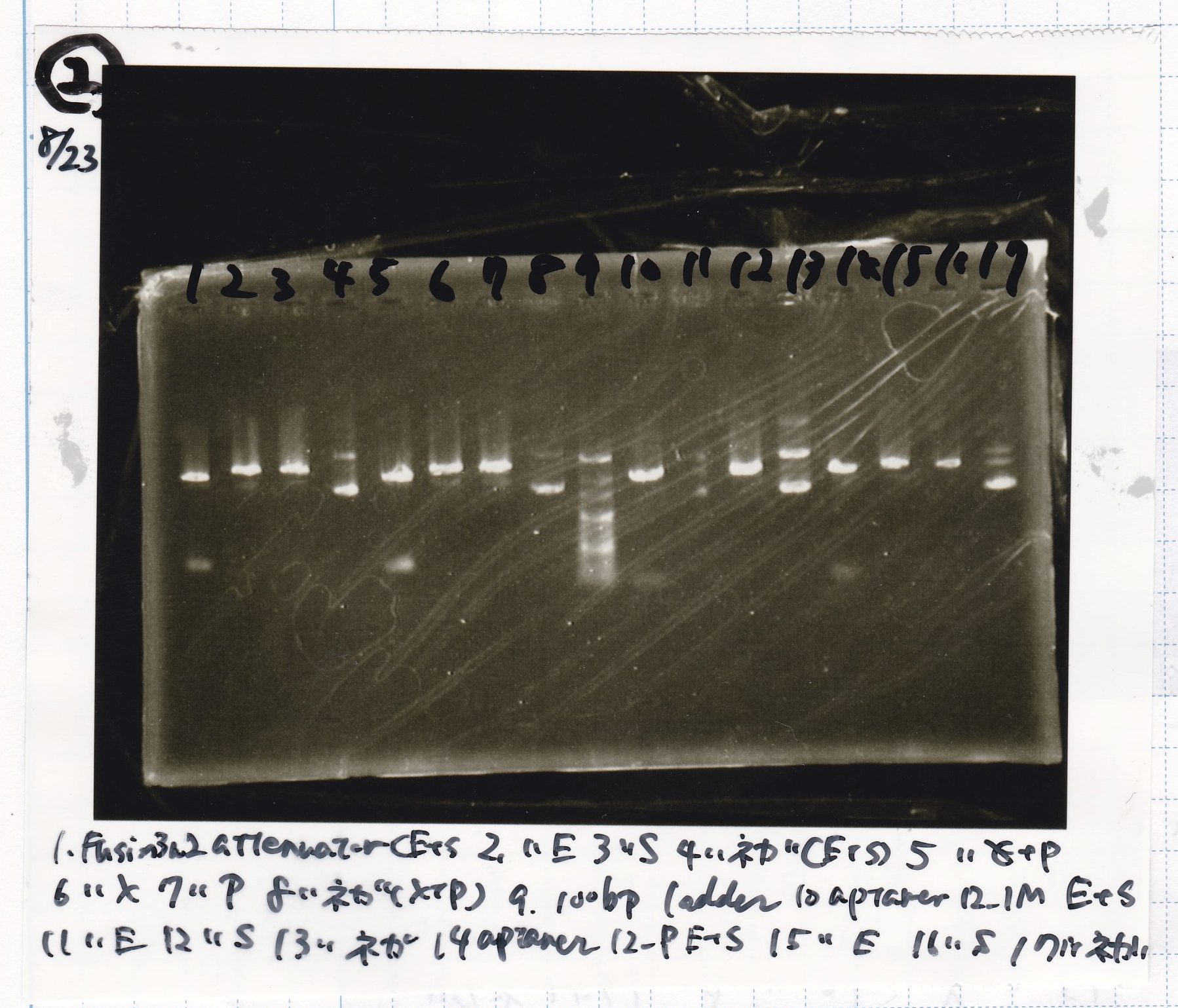
Kojima and Honda
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | Plac | SpeI | PstI
|
| 2 | Plac | SpeI | --
|
| 3 | Plac | -- | PstI
|
| 4 | Plac | -- | --
|
| 5 | Spinach | EcoRI | SpeI
|
| 6 | Spinach | EcoRI | --
|
| 7 | Spinach | -- | SpeI
|
| 8 | Spinach | -- | --
|
| 9 | 100bp ladder | -- | --
|
| 10 | J23100 | SpeI | PstI
|
| 11 | J23100 | SpeI | --
|
| 12 | J23100 | -- | PstI
|
| 13 | J23100 | -- | --
|
| 14 | tetR aptamer 12_1R | EcoRI | SpeI
|
| 15 | tetR aptamer 12_1R | EcoRI | --
|
| 16 | tetR aptamer 12_1R | -- | SpeI
|
| 17 | tetR aptamer 12_1R | -- | --
|
Restriction Enzyme Digestion
Nakamoto and Kojima
| | Fusion6 antisense (1) | EcoRI | SpeI | buffer B | BSA | MilliQ | total
|
| 2 cuts | 3 | 0.5 | 0.5 | 3 | 0.3 | 22.7 | 30
|
| 1 cut | 0.6 | 0.1 | 0 | 1 | 0.1 | 8.2 | 10
|
| 1 cut | 0.6 | 0 | 0.1 | 1 | 0.1 | 8.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 0.1 | 8.3 | 10
|
| | Fusion6 antisense (1) | XbaI | PstI | buffer D | BSA | MilliQ | total
|
| 2 cuts | 3 | 0.5 | 0.5 | 3 | 0.3 | 22.7 | 30
|
| 1 cut | 0.6 | 0.1 | 0 | 1 | 0.1 | 8.2 | 10
|
| 1 cut | 0.6 | 0 | 0.1 | 1 | 0.1 | 8.2 | 10
|
| NC | 0.6 | 0 | 0 | 1 | 0.1 | 8.3 | 10
|
| | Fusion1 antisense (1) | EcoRI | SpeI | buffer B | BSA | MilliQ | total
|
| 2 cuts | 3.3 | 0.5 | 0.5 | 3 | 0.3 | 22.4 | 30
|
| 1 cut | 0.7 | 0.1 | 0 | 1 | 0.1 | 8.1 | 10
|
| 1 cut | 0.7 | 0 | 0.1 | 1 | 0.1 | 8.1 | 10
|
| NC | 0.7 | 0 | 0 | 1 | 0.1 | 8.2 | 10
|
| | pT181 antisense (1) | EcoRI | SpeI | buffer B | BSA | MilliQ | total
|
| 2 cuts | 5.6 | 0.5 | 0.5 | 3 | 0.3 | 20.1 | 30
|
| 1 cut | 1.1 | 0.1 | 0 | 1 | 0.1 | 7.7 | 10
|
| 1 cut | 1.1 | 0 | 0.1 | 1 | 0.1 | 7.7 | 10
|
| NC | 1.1 | 0 | 0 | 1 | 0.1 | 7.8 | 10
|
| | pT181 antisense (1) | XbaI | PstI | buffer D | BSA | MilliQ | total
|
| 2 cuts | 5.6 | 0.5 | 0.5 | 3 | 0.3 | 20.1 | 30
|
| 1 cut | 1.1 | 0.1 | 0 | 1 | 0.1 | 7.7 | 10
|
| 1 cut | 1.1 | 0 | 0.1 | 1 | 0.1 | 7.7 | 10
|
| NC | 1.1 | 0 | 0 | 1 | 0.1 | 7.8 | 10
|
- incubate at 37°C from 17:29.
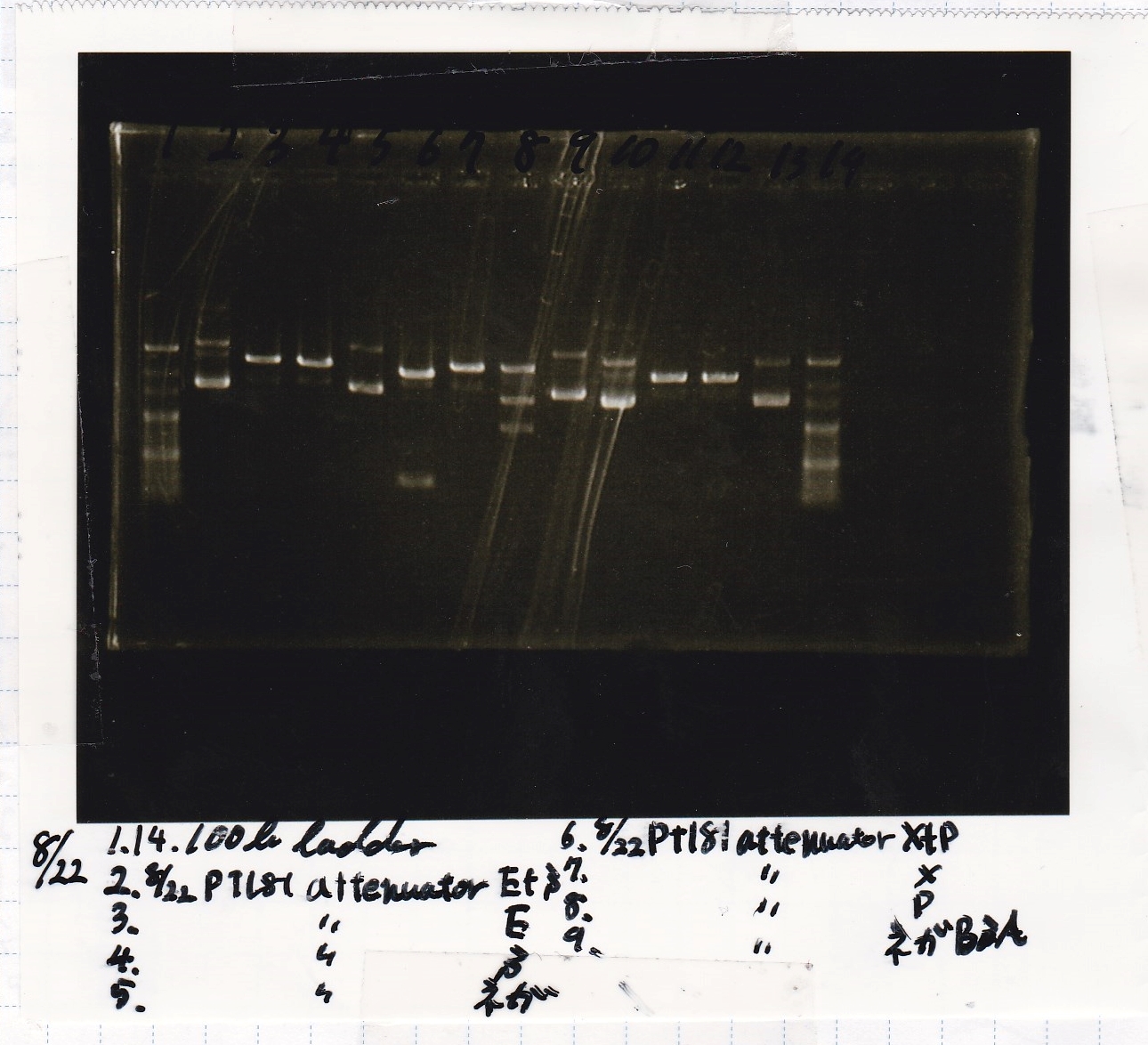
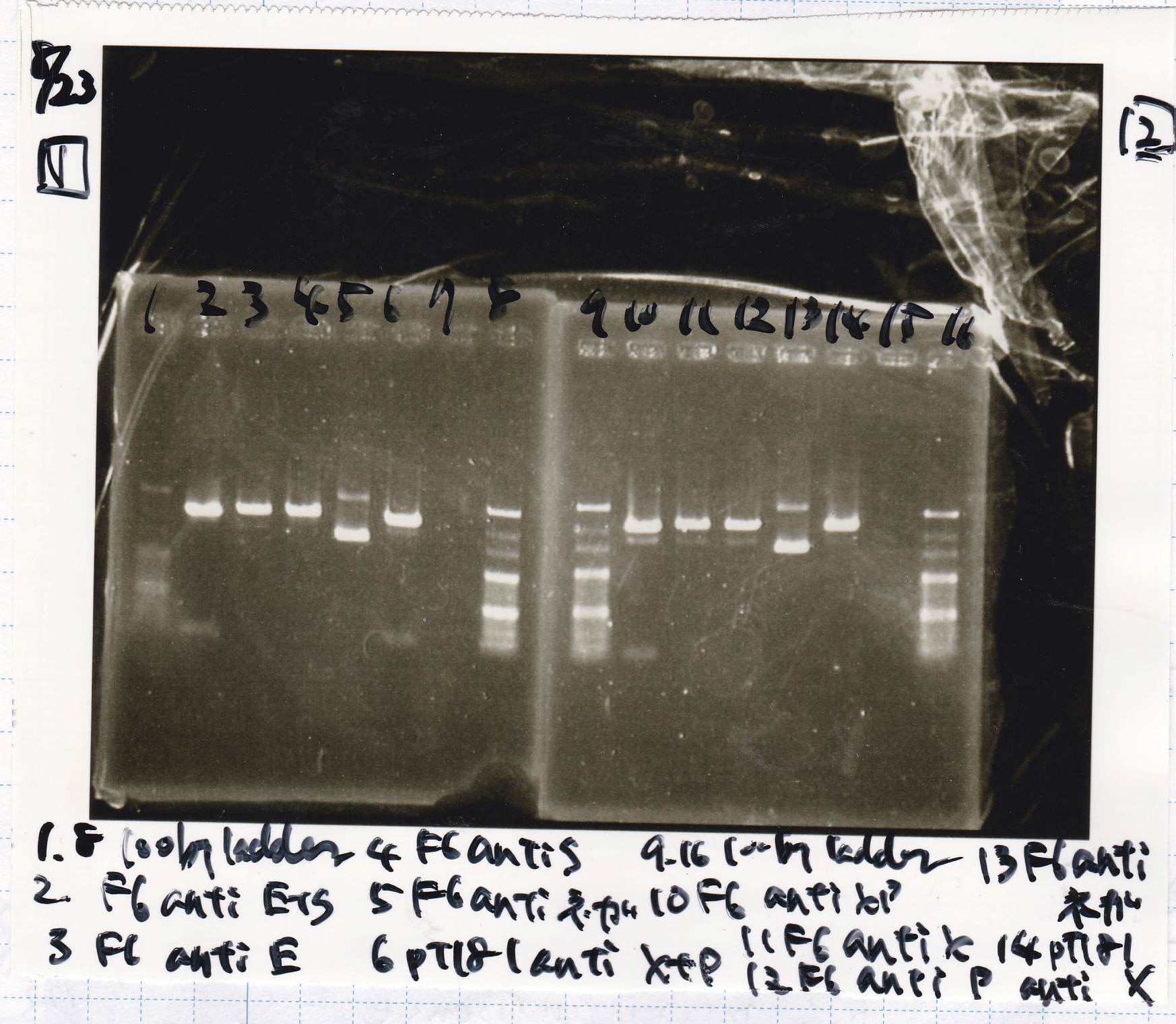
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp | -- | --
|
| 2 | pT181 attenuator | EcoRI | SpeI
|
| 3 | pT181 attenuator | XbaI | PstI
|
| 4 | Fusion3m2 attenuator | EcoRI | SpeI
|
| 5 | Fusion1 attenuator | XbaI | PstI
|
| 6 | Fusion1 attenuator | EcoRI | SpeI
|
| 7 | Fusion3m2 attenuator | XbaI | PstI
|
| 8 | pT181 antisense | EcoRI | SpeI
|
| 9 | pT181 antisense | XbaI | PstI
|
| 10 | Fusion1 antisense | EcoRI | SpeI
|
| 11 | Fusion1 antisense | XbaI | PstI
|
| 12 | Fusion6 antisense | EcoRI | SpeI
|
| 13 | Fusion6 antisense | XbaI | PstI
|
| 14 | tetR aptamer 12_1M | EcoRI | SpeI
|
| 15 | tetR aptamer 12_P | EcoRI | SpeI
|
| 16 | tetR aptamer 12_1R | EcoRI | SpeI
|
| 17 | 100bp | -- | --
|

| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp | -- | --
|
| 2 | Spinach | EcoRI | SpeI
|
| 3 | J23100 | SpeI | PstI
|
| 4 | Plac | SpeI | PstI
|
| 5 | 100bp | -- | --
|
| 6 | J23100 | SpeI | PstI
|
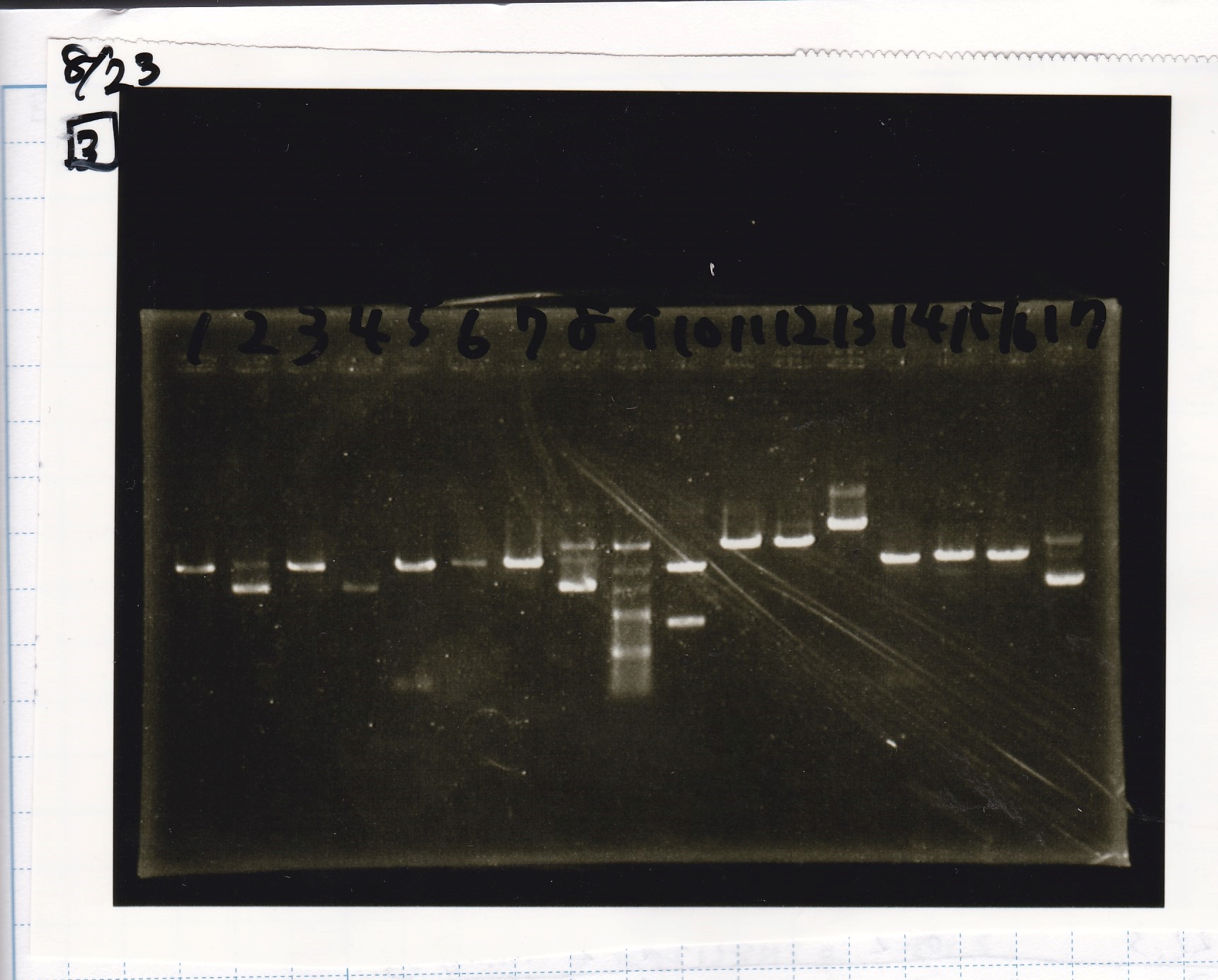
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp | -- | --
|
| 2 | Fusion6 antisense | EcoRI | SpeI
|
| 3 | Fusion6 antisense | EcoRI | --
|
| 4 | Fusion6 antisense | -- | SpeI
|
| 5 | Fusion6 antisense | -- | --
|
| 6 | pT181 antisense | XbaI | PstI
|
| 8 | 100bp | -- | --
|
| 9 | 100bp | -- | --
|
| 10 | Fusion6 antisense | XbaI | PstI
|
| 11 | Fusion6 antisense | XbaI | --
|
| 12 | Fusion6 antisense | -- | PstI
|
| 13 | Fusion6 antisense | -- | --
|
| 14 | pT181 antisense | XbaI | --
|
| 16 | 100bp | -- | --
|
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp | -- | --
|
| 2 | Fusion1 antisense | EcoRI | SpeI
|
| 3 | Fusion1 antisense | EcoRI | --
|
| 4 | Fusion1 antisense | -- | SpeI
|
| 5 | Fusion1 antisense | -- | --
|
| 6 | pT181 antisense | -- | PstI
|
| 8 | 100bp | -- | --
|
| 9 | 100bp | -- | --
|
| 10 | pT181 antisense | EcoRI | SpeI
|
| 11 | pT181 antisense | EcoRI | --
|
| 12 | pT181 antisense | -- | SpeI
|
| 13 | pT181 antisense | -- | --
|
| 14 | pT181 antisense | -- | --
|
| 16 | 100bp | -- | --
|

Nakamoto, Ashida, Kojima, Okazaki & Matsumoto
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | pT181 attenuator | XbaI & PstI
|
| 3
|
| 5 | Fusion3m2 attenuator | EcoRI & SpeI
|
| 6
|
| 8 | Fusion1 attenuator | XbaI & PstI
|
| 9
|
| 11 | Fusion1 attenuator | EcoRI & SpeI
|
| 12
|

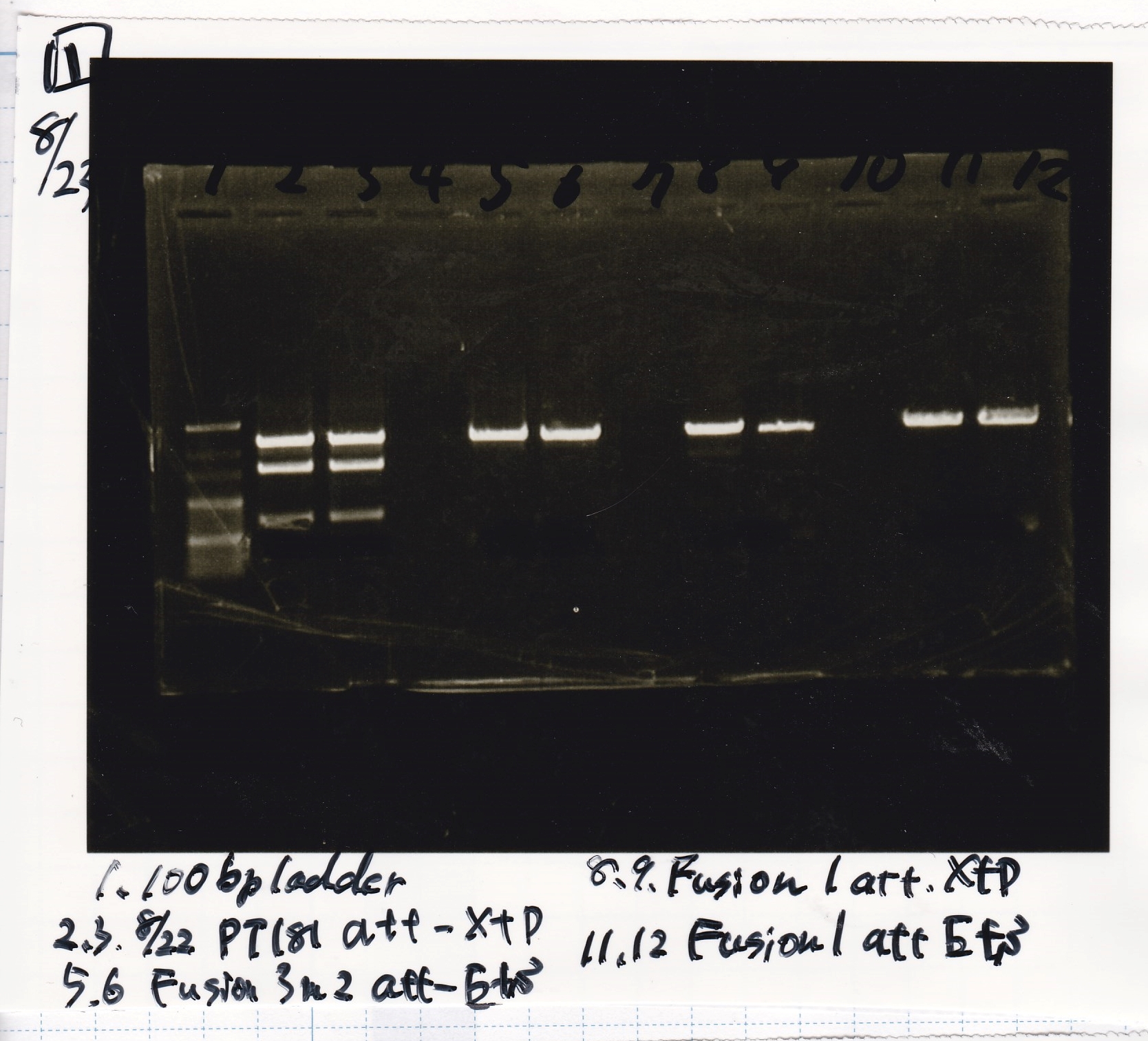
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | Fusion3m2 attenuator | XbaI & PstI
|
| 3
|
| 5 | pT181 antisense | EcoRI & SpeI
|
| 6
|
| 8 | pT181 antisense | XbaI & PstI
|
| 9
|
| 11 | Fusion1 antisense | XbaI & PstI
|
| 12
|

| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | Fusion6 attenuator | XbaI & PstI
|
| 3
|
| 5 | aptamer 12_1M | EcoRI & SpeI
|
| 6
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | aptamer 12_P | EcoRI & SpeI
|
| 3
|
| 5 | aptamer 12_1R | EcoRI & SpeI
|
| 6
|
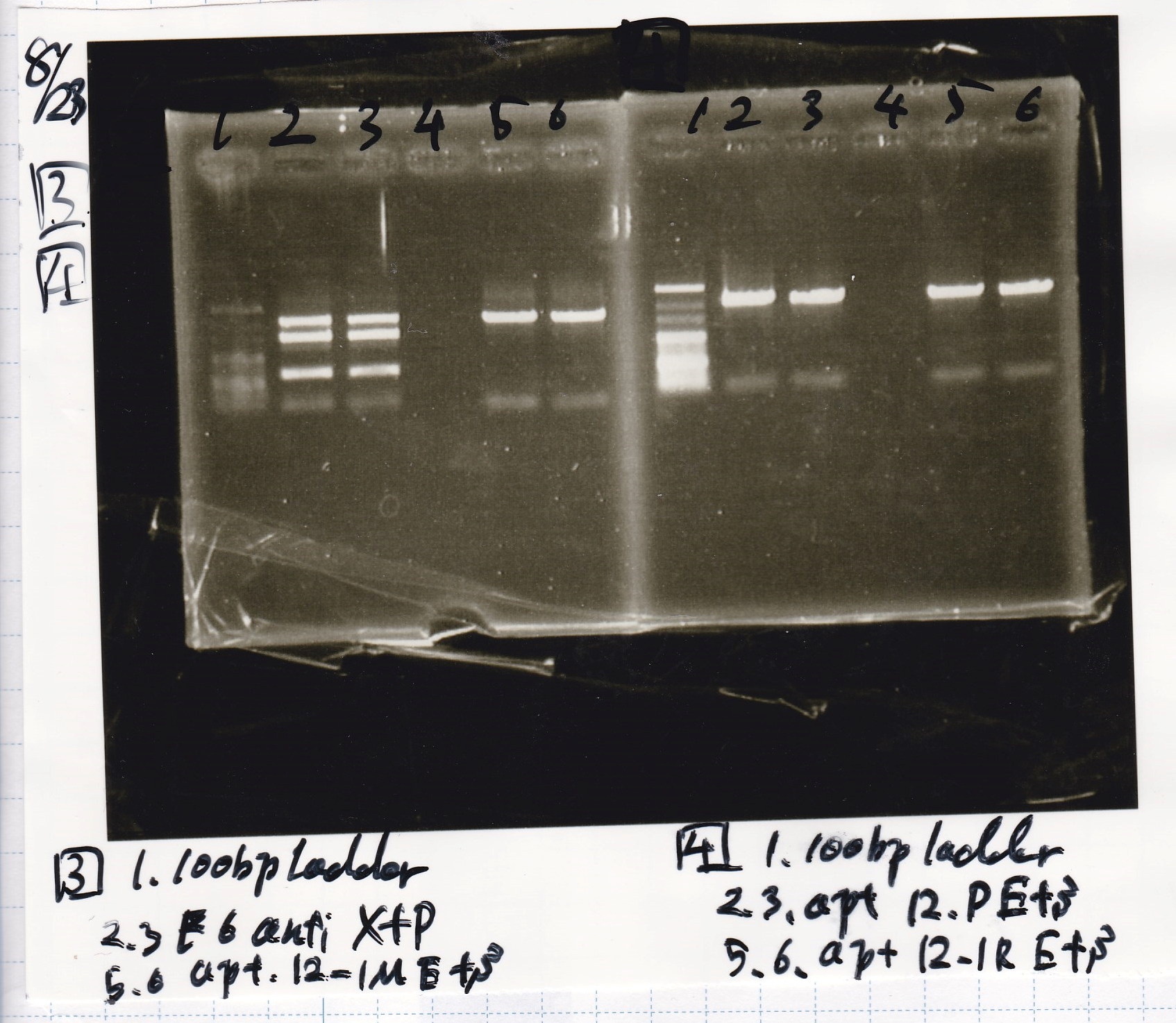
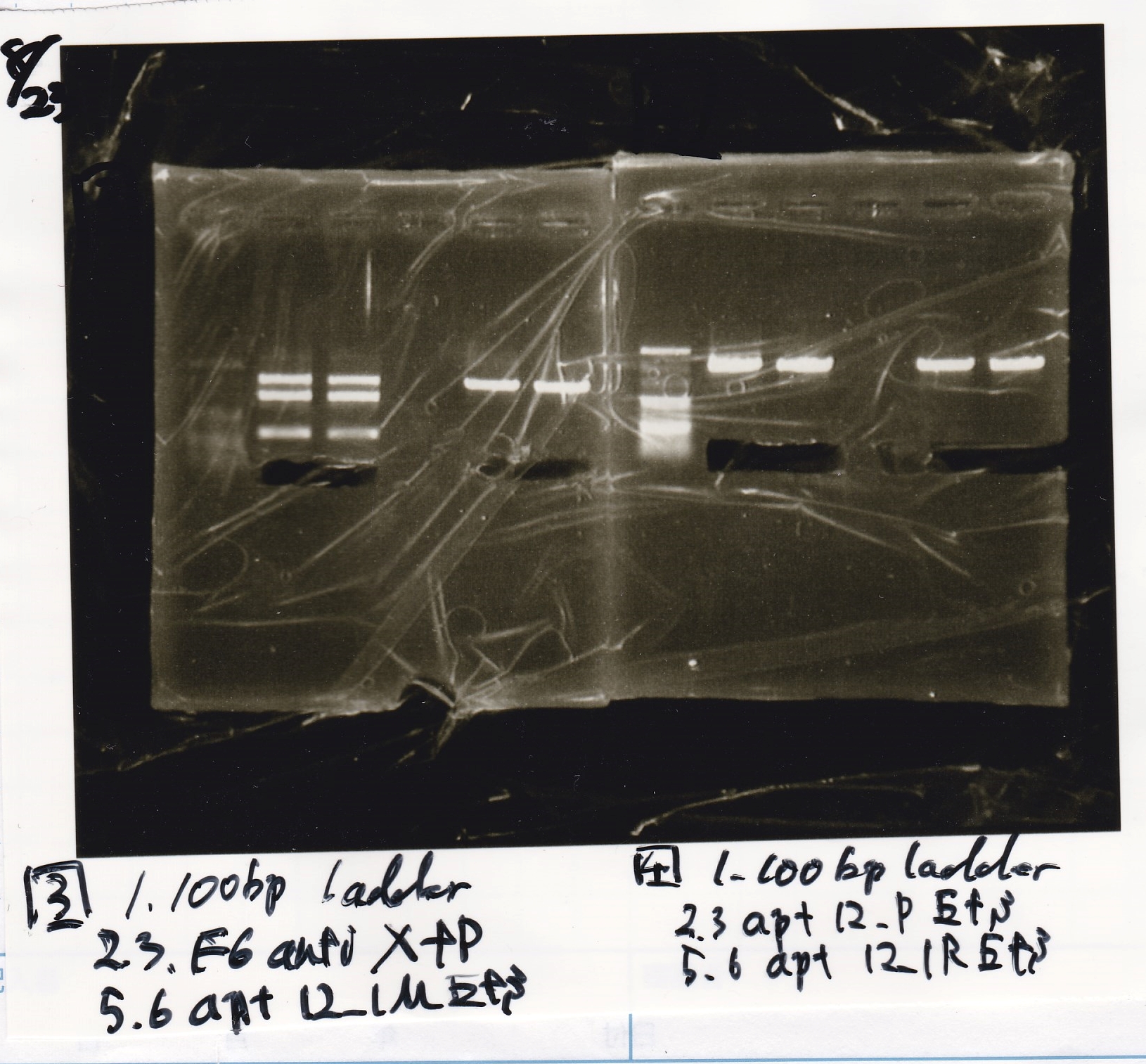
Ashida
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | Fusion6 antisense | EcoRI & SpeI
|
| 3
|
| 5 | Fusion6 attenuator | XbaI & PstI
|
| 6
|
| 8 | Fusion1 antisense | EcoRI & SpeI
|
| 9
|
| 11 | pT181 antisense | EcoRI & SpeI
|
| 12
|

| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 3 | pT181 antisense | XbaI & PstI
|
| 4
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 3 | Plac | SpeI & PstI
|
| 4
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 3 | J23100 | SpeI & PstI
|
| 5 | Spinach | EcoRI & SpeI
|
| 6
|


Aug 24
Ashida
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/22 aptamer 12_P (EcoRI & SpeI) | 88 | 21.3 | --
|
| 8/22 aptamer 12_1R (EcoRI & SpeI) | 63 | 23.1 | --
|
| 8/22 aptamer 12_1M (EcoRI & SpeI) | 40 | 22.0 | --
|
| 8/23 Fusion6 antisense (EcoRI & SpeI) | 6.0 | 1.60 | 0.17
|
| 8/23 Fusion6 antisense (XbaI & PstI) | 6.0 | 1.71 | 0.25
|
| 8/23 pT181 antisense (EcoRI & SpeI) | 6.0 | 2.22 | 0.15
|
| 8/23 pT181 antisense (XbaI & PstI) | 6.0 | 2.10 | 0.04
|
| 8/22 Plac (SpeI & PstI) | 6.0 | 1.47 | 0.09
|
| 8/22 Fusion1 antisense (EcoRI & SpeI) | 10 | 2.16 | 0.01
|
| 8/22 J23100 (SpeI & PstI) | 6.0 | 1.52 | 0.35
|
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/22 Spinach (EcoRI & SpeI) | 8.0 | 1.15 | 0.77
|
| 8/22 Fusion6 antisense (XbaI & PstI) | 44 | 1.07 | 0.96
|
| 8/22 Fusion1 antisense (XbaI & PstI) | 2 | -- | 1.00
|
| 8/22 pT181 antisense (XbaI & PstI) | 3. | 2.01 | 0.40
|
| 8/22 pT181 antisense (EcoRI & SpeI) | 8 | 1.13 | 0.82
|
| 8/22 Fusion3m2 attenuator (XbaI & PstI) | 5 | 2.27 | 1.50
|
| 8/22 Fusion3m2 attenuator (EcoRI & SpeI) | 34 | 1.16 | 1.02
|
| 8/22 Fusion1 attenuator (XbaI & PstI) | 14 | 1.40 | 0.62
|
| 8/22 Fusion1 attenuator (EcoRI & SpeI) | 16 | 1.27 | 0.61
|
| 8/22 pT181 attenuator (XbaI & PstI) | 5 | -- | 0.88
|
- These samples were performed ethanol ethanl precipitation.
Restriction Enzyme Digestion
No name
| | tetR aptamer 12_P(2) | EcoRI | SpeI | 10x Buffer B | 100x BSA | MilliQ | total
|
| 2 cuts | 2.7µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 23µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 0.1µL | 8.6µL | 10µL
|
| | tetR aptamer 12_1R(2) | EcoRI | SpeI | 10x Buffer B | 100x BSA | MilliQ | total
|
| 2 cuts | 2.6µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 23.1µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 0.1µL | 8.6µL | 10µL
|
| | tetR aptamer 12_M(1) | EcoRI | SpeI | 10x Buffer B | 100x BSA | MilliQ | total
|
| 2 cuts | 2.7µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 23µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 0.1µL | 8.6µL | 10µL
|
Master Plate
No name
| Number | Use LB plate(+Kan)
|
| 1 | entA- #1
|
| 2 | entA- #1
|
| 3 | entA- #1
|
| 4 | entA- #2
|
| 5 | entA- #2
|
| 6 | entA- #2
|
Electrophoresis
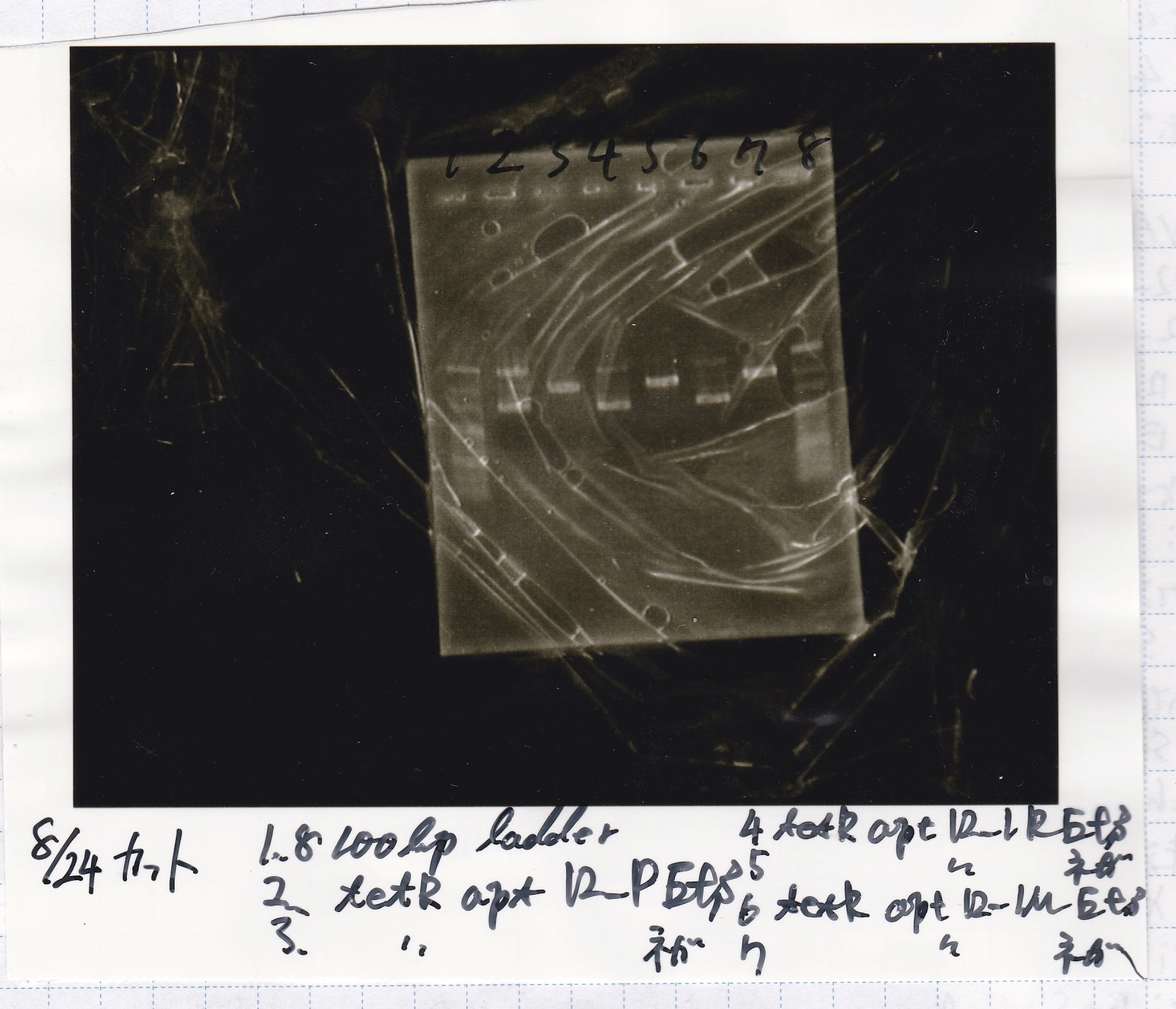
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | - | -
|
| 2 | tetR aptamer 12_P | EcoRI | SpeI
|
| 3 | tetR aptamer 12_P | - | -
|
| 4 | tetR aptamer 12_1R | EcoRI | SpeI
|
| 5 | tetR aptamer 12_1R | - | -
|
| 6 | tetR aptamer 12_1M | EcoRI | SpeI
|
| 7 | tetR aptamer 12_1M | - | -
|
| 8 | 100bp ladder | - | -
|
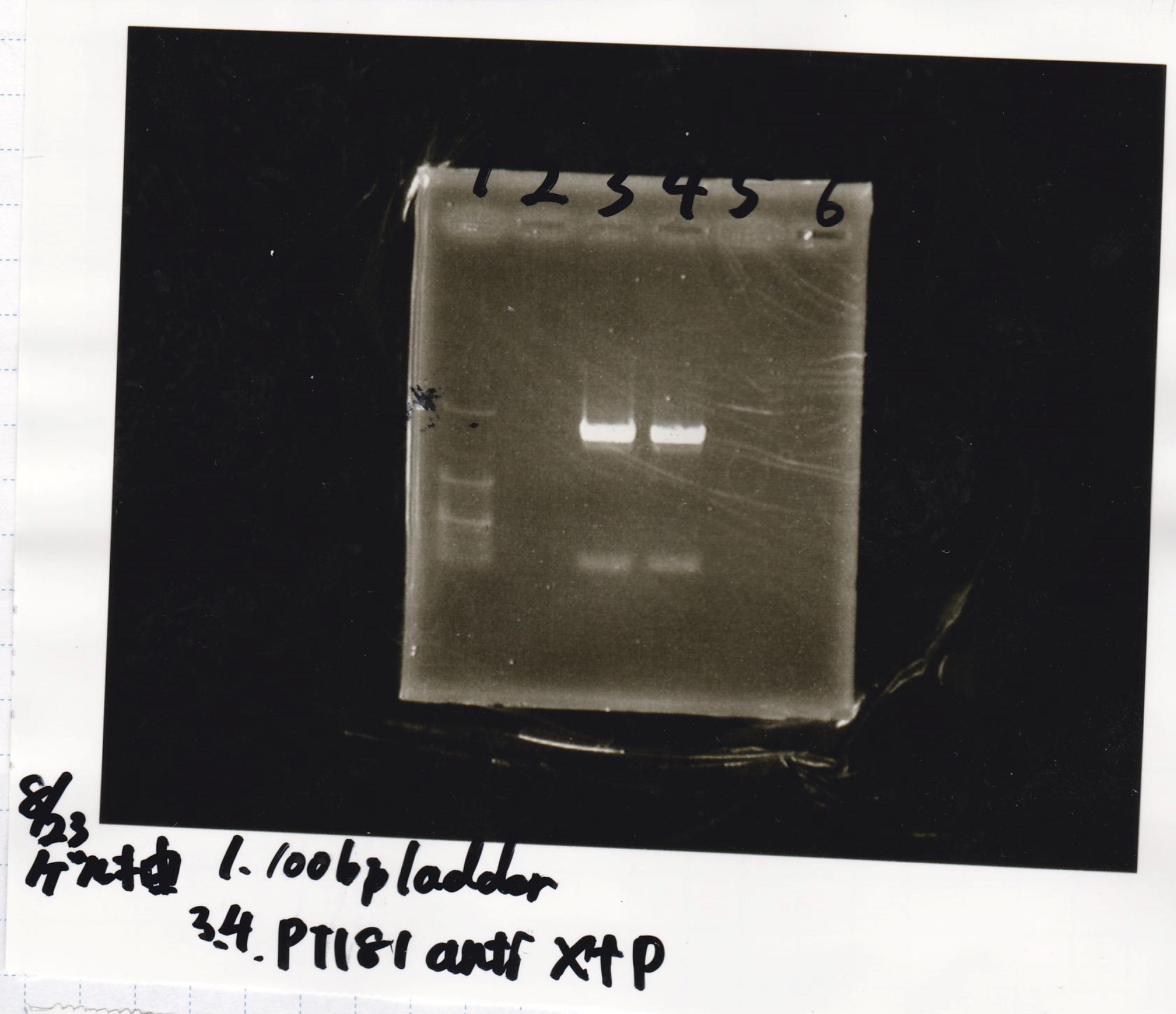
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | - | -
|
| 2 | tetR aptamer 12_P | EcoRI | SpeI
|
| 3 | tetR aptamer 12_1R | EcoRI | SpeI
|
| 4 | tetR aptamer 12_1M | EcoRI | SpeI
|
| 5 | 100bp ladder | - | -
|
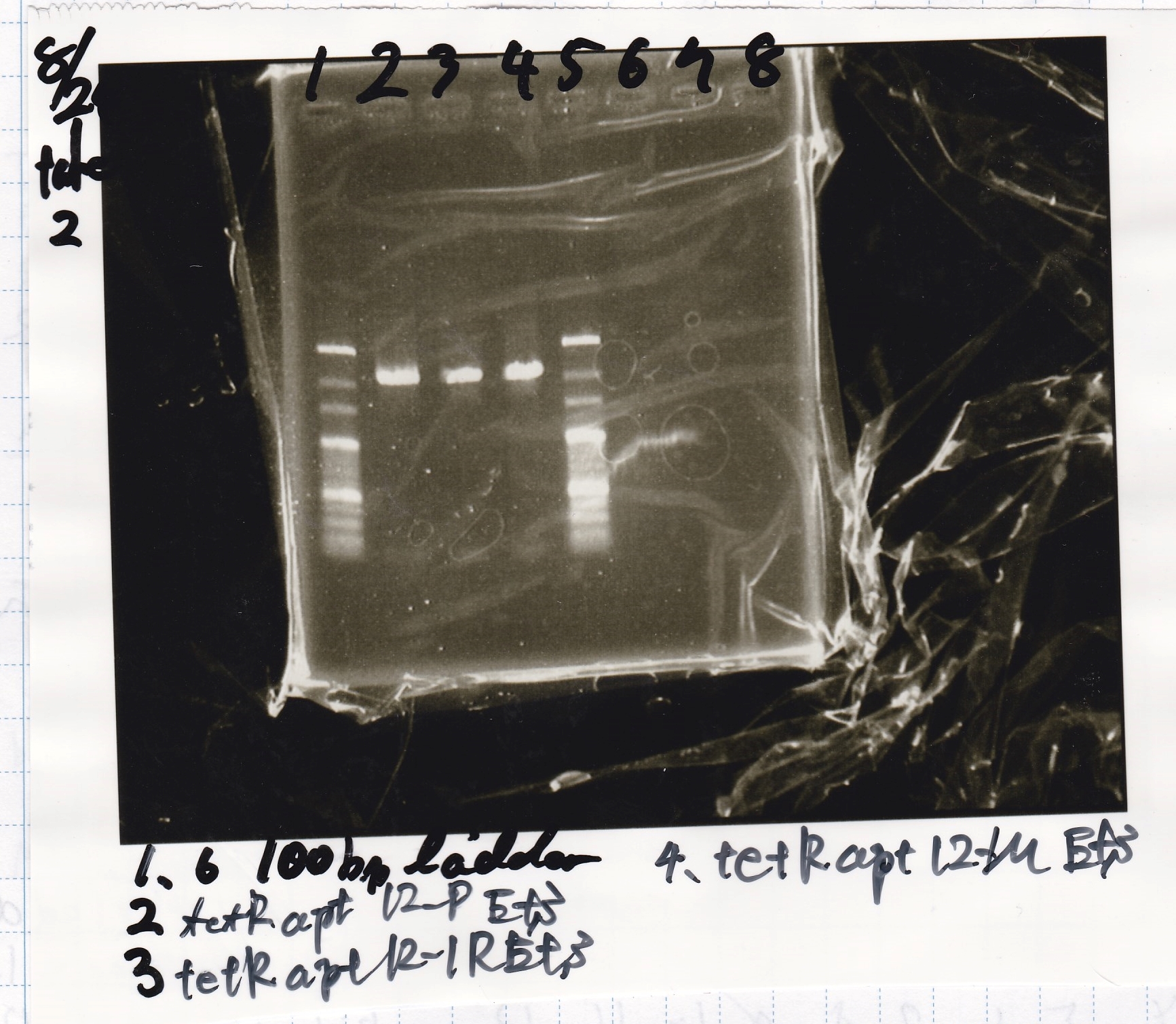
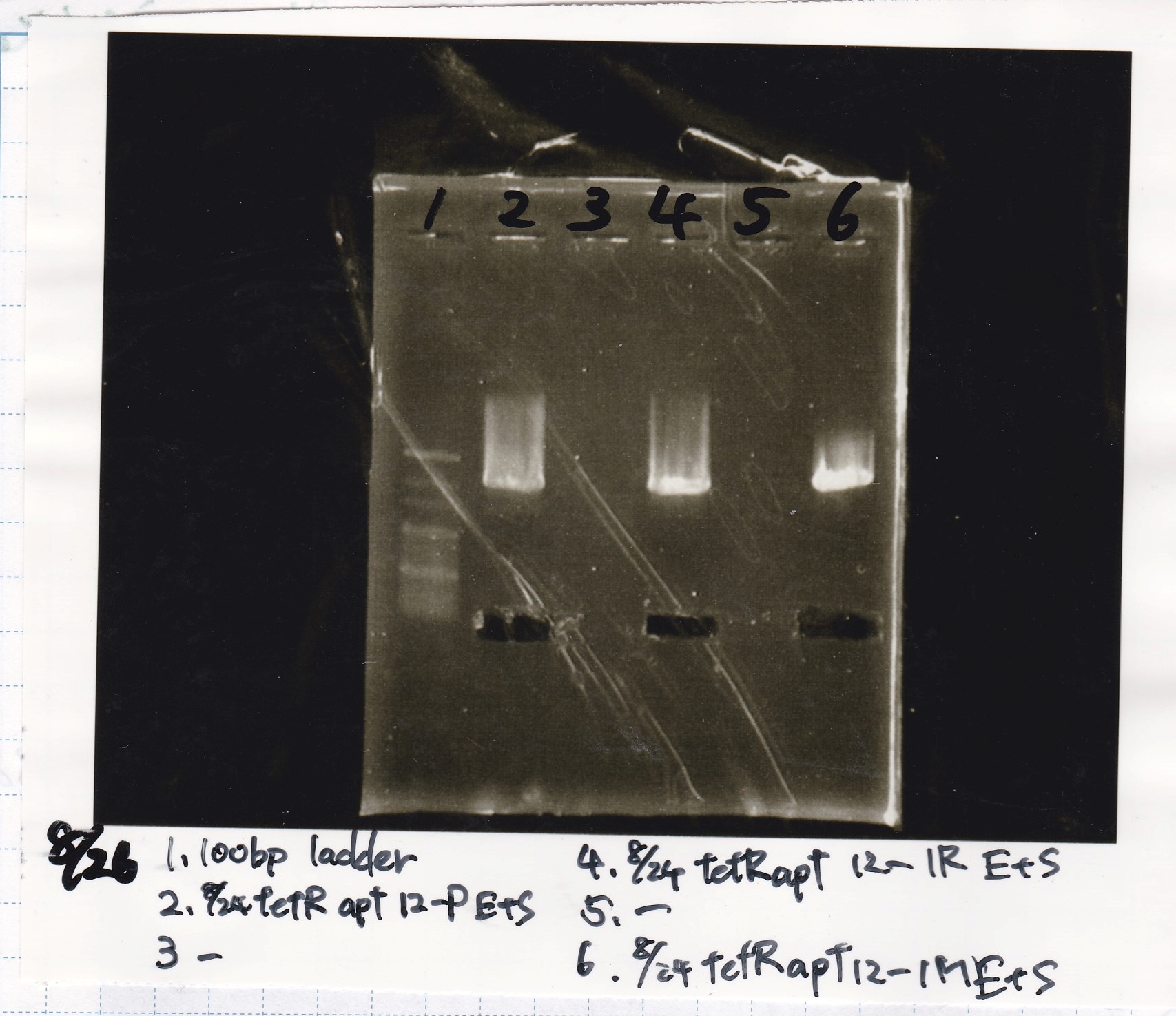
Aug 26
Matsumoto and Ashida
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | -
|
| 2 | 8/24 tetR aptamer 12_P | EcoRI & SpeI
|
| 4 | 8/24 tetR aptamer 12_1R | EcoRI & SpeI
|
| 6 | 8/24 tetR aptamer 12_M | EcoRI & SpeI
|
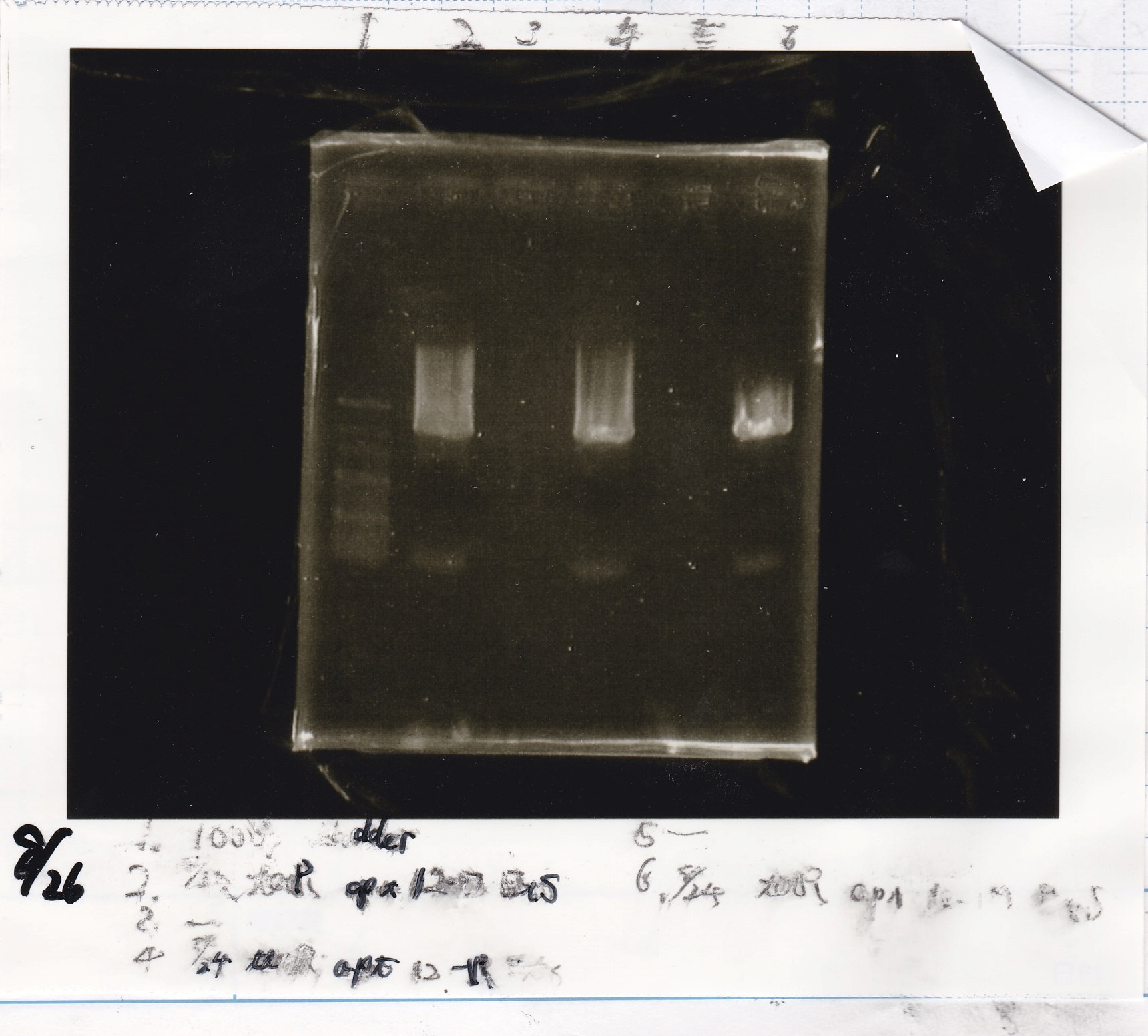
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| tetR aptamer 12_P (EcoRI & SpeI) | 2.3 | 0.38 | 0.13
|
| tetR aptamer 12_1R (EcoRI & SpeI) | 12.0 | 0.63 | 1.19
|
| tetR aptamer 12_M (EcoRI & SpeI) | 2.0 | 6.43 | 0.03
|
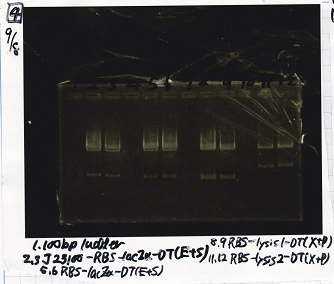
Ligation
Ashida
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 8/21 DT | 5.0 | 8/26 tetR aptamer 12_1R | 2.0 | 3.5
|
| experiment | 8/21 DT | 5.0 | 8/22 pT181 atteniator | 5.0 | 5.0
|
| experiment | 8/21 DT | 5.0 | 8/22 pT181 antisense | 3.0 | 4.0
|
| experiment | 8/21 DT | 5.0 | 8/22 Spinach | 3.0 | 4.0
|
| experiment | 8/22 Plac | 4.0 | 8/22 pT181 attenuator | 5.0 | 4.5
|
| experiment | 8/22 J23100 | 4.0 | 8/22 pT181 attenuator | 5.0 | 4.5
|
| experiment | 8/22 Plac | 4.0 | 8/22 pT181 antisense | 8.0 | 6.0
|
| experiment | 8/22 J23100 | 4.0 | 8/22 pT181 antisense | 8.0 | 6.0
|
Transformation
No name
| Name | Sample | Competent Cells | Total | Plate
|
| 8/26 tetR aptamer 12_1R-DT | 1µL | 10µL | 11µL | CP
|
| 8/26 pT181 attenuator-DT | 1µL | 10µL | 11µL | CP
|
| 8/26 pT181 antisense-DT | 1µL | 10µL | 11µL | CP
|
| 8/26 Spinach-DT | 1µL | 10µL | 11µL | CP
|
| 8/26 Plac-pT181 attenuator | 1µL | 10µL | 11µL | CP
|
| 8/26 Pcon-pT181 attenuator | 1µL | 10µL | 11µL | Amp
|
| 8/26 Plac-pT181 antisense | 1µL | 10µL | 11µL | CP
|
| 8/26 Pcon-pT181 antisense | 1µL | 10µL | 11µL | Amp
|
| Pλ-RBS-luxI-DT | 1µL | 10µL | 11µ | Amp
|
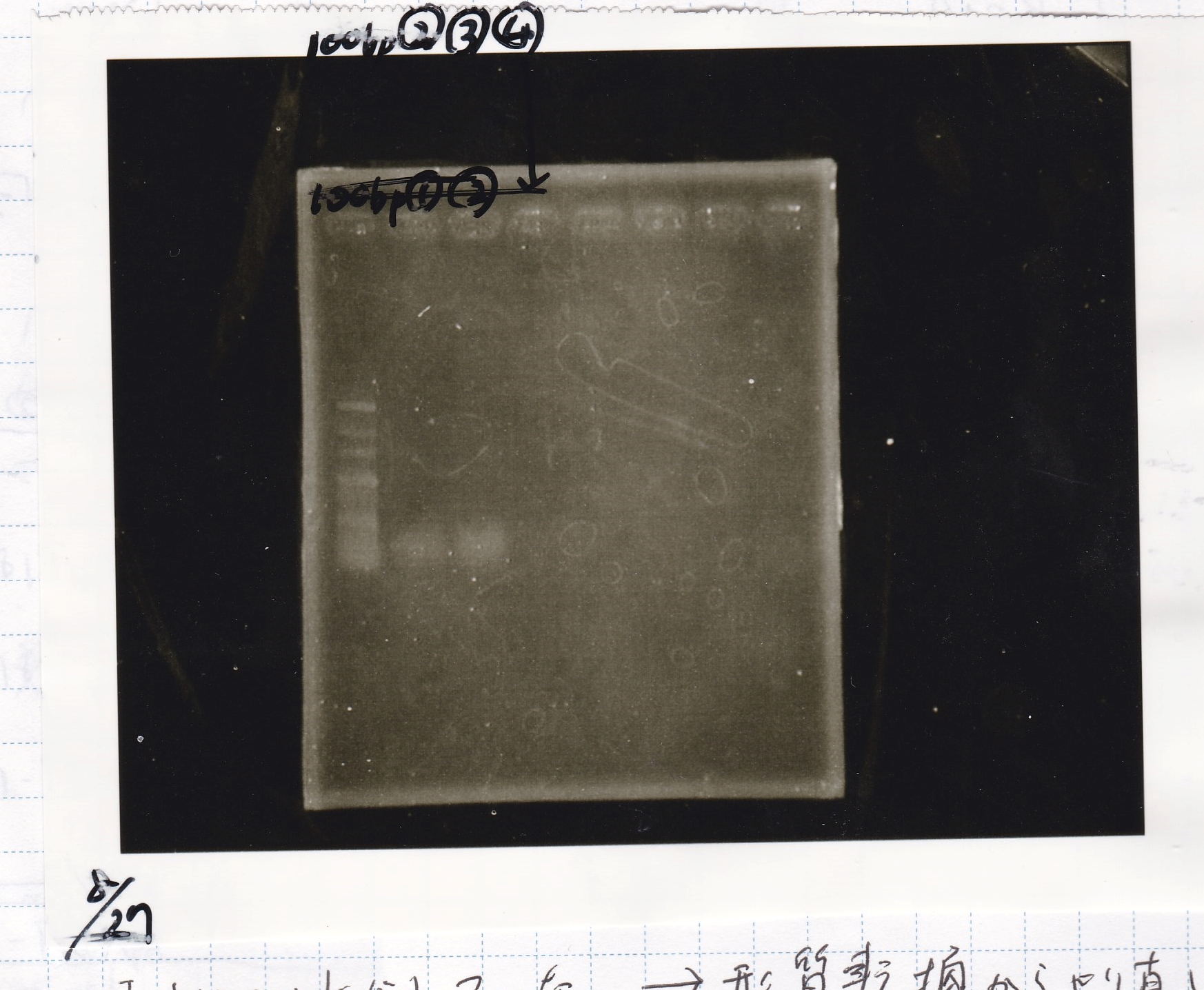
Aug 27
Restriction Enzyme Digestion
No name
| | 8/21 pT181 attenuator-1(330µg/mL) | EcoRI | SpeI | 10x BufferB | 100x BSA | MilliQ | total
|
| 2 cuts | 3.0µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.7µL | 30µL
|
| NC | 0.6µL | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL
|
| | 8/17 DT-1(188µg/mL) | EcoRI | XbaI | 10x BufferB | 100x BSA | MilliQ | total
|
| 2 cuts | 3.0µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.7µL | 30µL
|
| NC | 1.0µl | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
| | 8/20 Pcon-RBS-luxR-DT-2(344µg/mL) | EcoRI | SpeI | 10x BufferB | 100x BSA | MilliQ | total
|
| 2 cuts | 2.9µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.8µL | 30µL
|
| NC | 0.6µl | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | 8/21 pT181 attenuator | EcoI | SpeI
|
| 3 | 8/21 pT181 atteniator | -- | --
|
| 4 | 8/17 DT-1 | EcoRI | XbaI
|
| 5 | 8/17 DT-1 | -- | --
|
| 6 | 8/20 Pcon-RBS-luxR-DT-2 | EcoI | SpeI
|
| 7 | 8/20 Pcon-RBS-luxR-DT-2 | -- | --
|
| 8 | 100bp ladder | -- | --
|
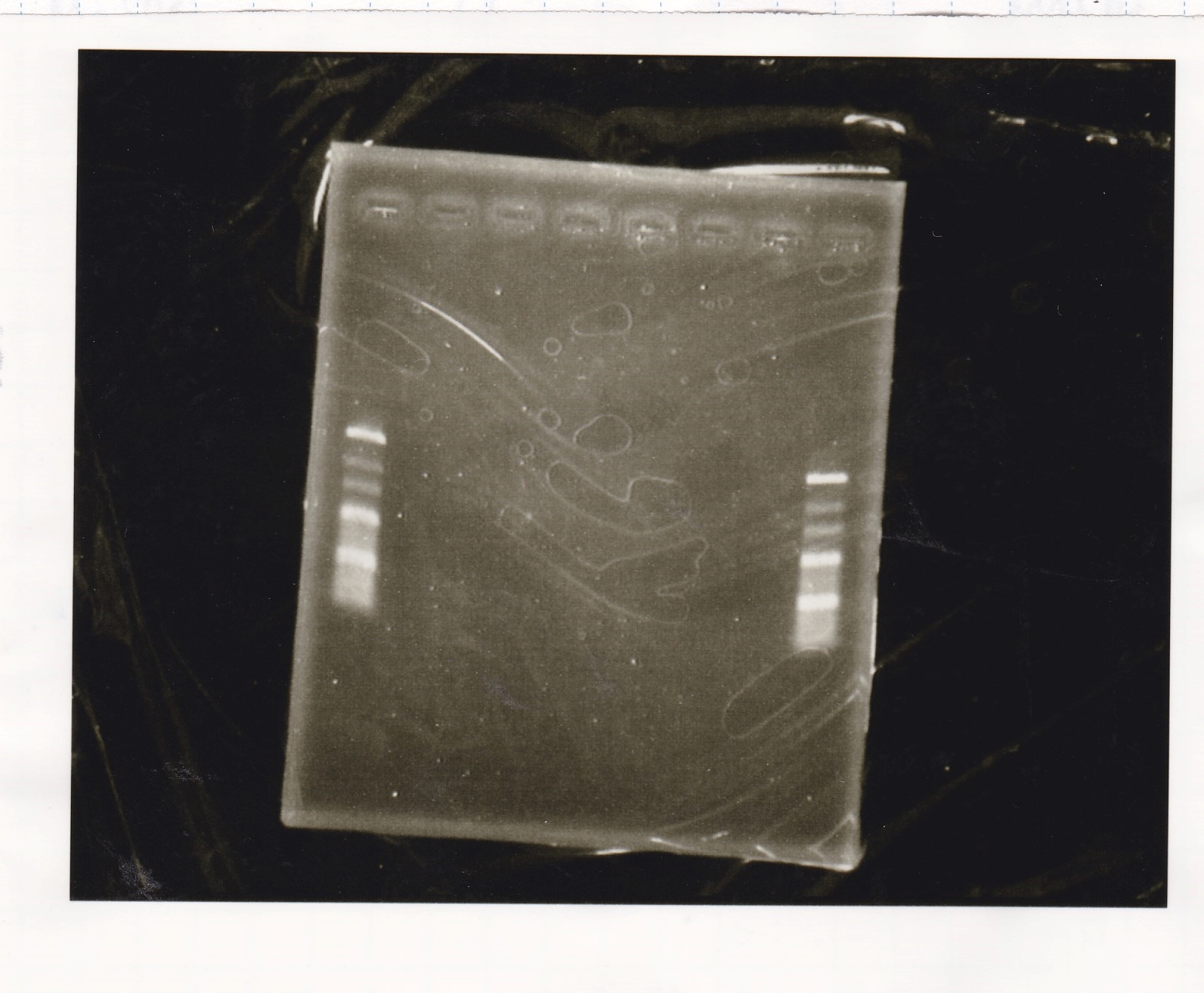
Colony PCR
Kojima and Yoshida
| Sample | base pair
|
| 8/26 Pλ-luxI(1) |
|
| 8/26 Pλ-luxI(2) |
|
| NC |
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 3min | 30cycles
|
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Pλ-luxI-(1)
|
| 3 | Pλ-luxI-(2)
|
| 4 | NC
|
Liquid Culture
No name
| Sample | medium
|
| 8/26 Pλ-RBS-luxI-DT-1 | Plusgrow medium (+Amp)
|
| 8/26 Pλ-RBS-luxI-DT-2 | Plusgrow medium (+Amp)
|
Restriction Enzyme Digestion
No name
| | 8/21 pT181 attenuator-1(330µg/mL) | EcoRI | SpeI | 10x BufferB | 100x BSA | MilliQ | total
|
| 2 cuts | 3.0µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.7µL | 30µL
|
| NC | 0.6µL | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL
|
| | 8/17 DT-1(188µg/mL) | EcoRI | XbaI | 10x BufferB | 100x BSA | MilliQ | total
|
| 2 cuts | 3.0µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.7µL | 30µL
|
| NC | 1.0µl | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
| | 8/20 Pcon-RBS-luxR-DT-2(344µg/mL) | EcoRI | SpeI | 10x BufferB | 100x BSA | MilliQ | total
|
| 2 cuts | 2.9µL | 0.5µL | 0.5µL | 3.0µL | 0.3µL | 22.8µL | 30µL
|
| NC | 0.6µl | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL
|
Electrophoresis
Hirano
| Lane | Sample | Enzyme1 | Enzyme2
|
| 100bp ladder | -- | --
|
| 1 | pT181 attenuator | EcoRI | SpeI
|
| 2 | pT181 attenuator | -- | --
|
| 3 | DT-(1) | EcoRI | XbaI
|
| 4 | DT-(1) | -- | --
|
| 5 | Pcon-RBS-luxR-DT-(2) | EcoRI | SpeI
|
| 6 | Pcon-RBS-luxR-DT-(2) | -- | --
|
| 100bp ladder | -- | --
|
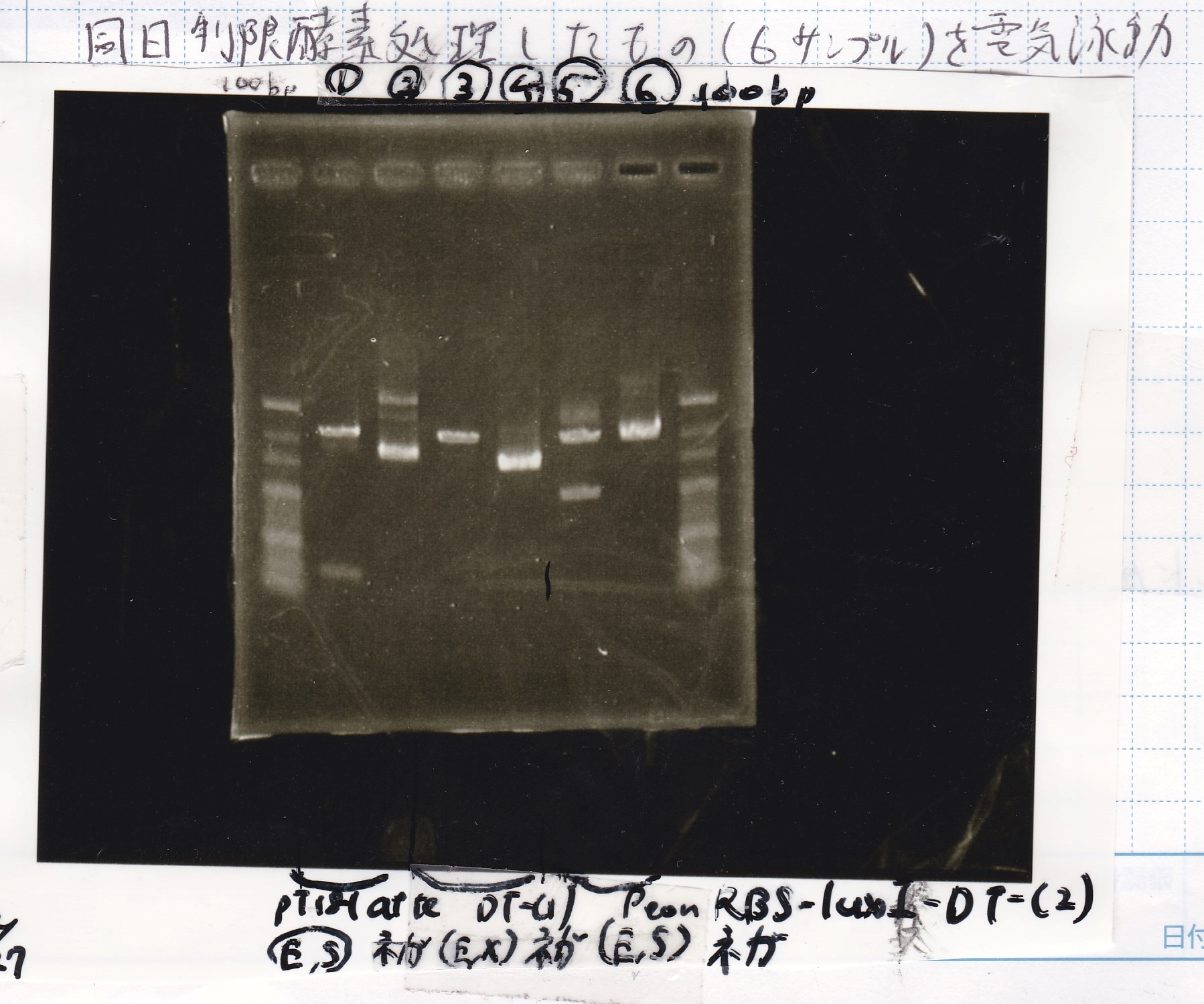
Inoue
| Lane | DNA | Enzyme
|
| 1 | 100bpladder | --
|
| 2 | pT181 attenuator-1 | EcoRI&SpeI
|
| 3 | pT181 attenuator-1 | EcoRI&SpeI
|
| 4 | DT-1 | EcoRI&XbaI
|
| 5 | DT-1 | EcoRI&XbaI
|
| 6 | Pcon-RBS-luxR-DT-2 | EcoRI&SpeI
|
| 7 | Pcon-RBS-luxR-DT-2 | EcoRI&SpeI
|
| 8 | 100bpladder | --
|

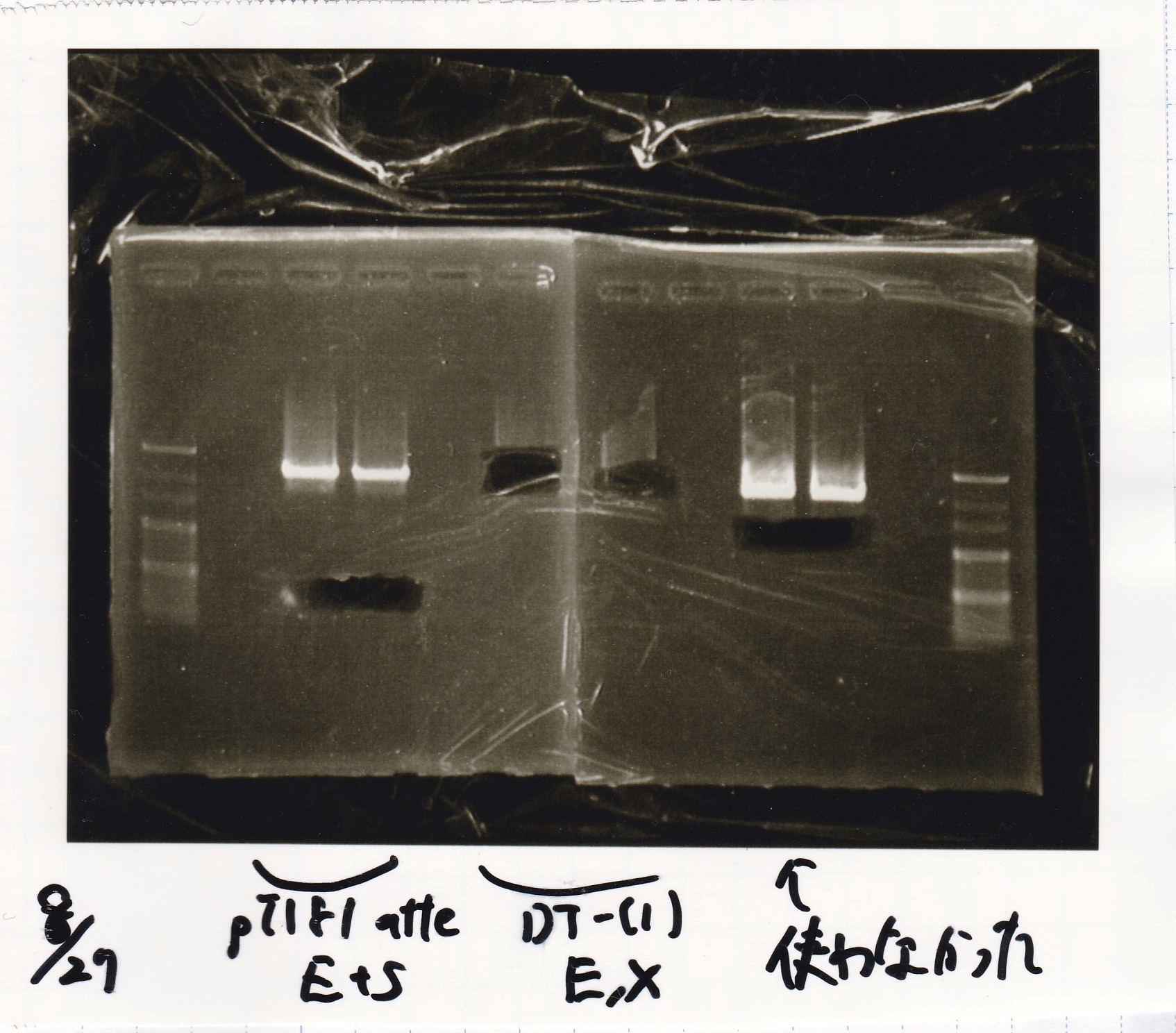
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| pT181 attenuator-1(EcoRI&SpeI) | 4.2 | 1.62 | 0.29
|
| DT-1(EcoRI&XbaI) | 17 | 1.88 | 0.88
|
Transformation
Kojima and Hirano
| Name | Sample | Competent Cells | Total | Plate
|
| 8/26 tetR aptamer 12_1R-DT | 1µL | 10µL | 11µL | CP
|
| 8/26 pT181 attenuator-DT | 1µL | 10µL | 11µL | CP
|
| 8/26 pT181 antisense-DT | 1µL | 10µL | 11µL | CP
|
| 8/26 Spinach-DT | 1µL | 10µL | 11µL | CP
|
| 8/26 Plac-pT181 attenuator | 1µL | 10µL | 11µL | CP
|
| 8/26 Pcon-pT181 attenuator | 1µL | 10µL | 11µL | Amp
|
| 8/26 Plac-pT181 antisense | 1µL | 10µL | 11µL | CP
|
| 8/26 Pcon-pT181 antisense | 1µL | 10µL | 11µL | Amp
|
Liquid culture
No name
| Sample | medium
|
| 8/16 Plux | Plusgrow medium(+CP)
|
Genome PCR
Yoshdia and Nakagawa
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | SasA_fwd primer | SasA_rev primer | MilliQ | total
|
| 0.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16 | 25
|
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | RpaA_fwd primer | RpaA_rev primer | MilliQ | total
|
| 0.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16 | 25
|
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | RpaB_fwd primer | RpaB_rev primer | MilliQ | total
|
| 0.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16 | 25
|
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | PkaiBC_fwd primer | PkaiBC_rev primer | MilliQ | total
|
| 0.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 50°C | 68°C | --
|
| 2 min | 10 sec | 30 sec | 38 sec | 30 cycles
|
Ligation
No name
| state | Vector | Inserter
|
| experiment | 8/27 DT (EcoRI & XbaI) | 2.8 | 8/21 RBS-lysis3 (EcoRI & SpeI)
|
| experiment | 8/27 DT (EcoRI & XbaI) | 2.8 | 8/27 pT181 attenuator (EcoRI & SpeI)
|
- Samples were evaporeted used evaporator into about 3 µL.
| sample | MilliQ | Ligation High | total
|
| 3 | 4 | 3.5 | 10.5
|
- incubate overnight at 4 °C
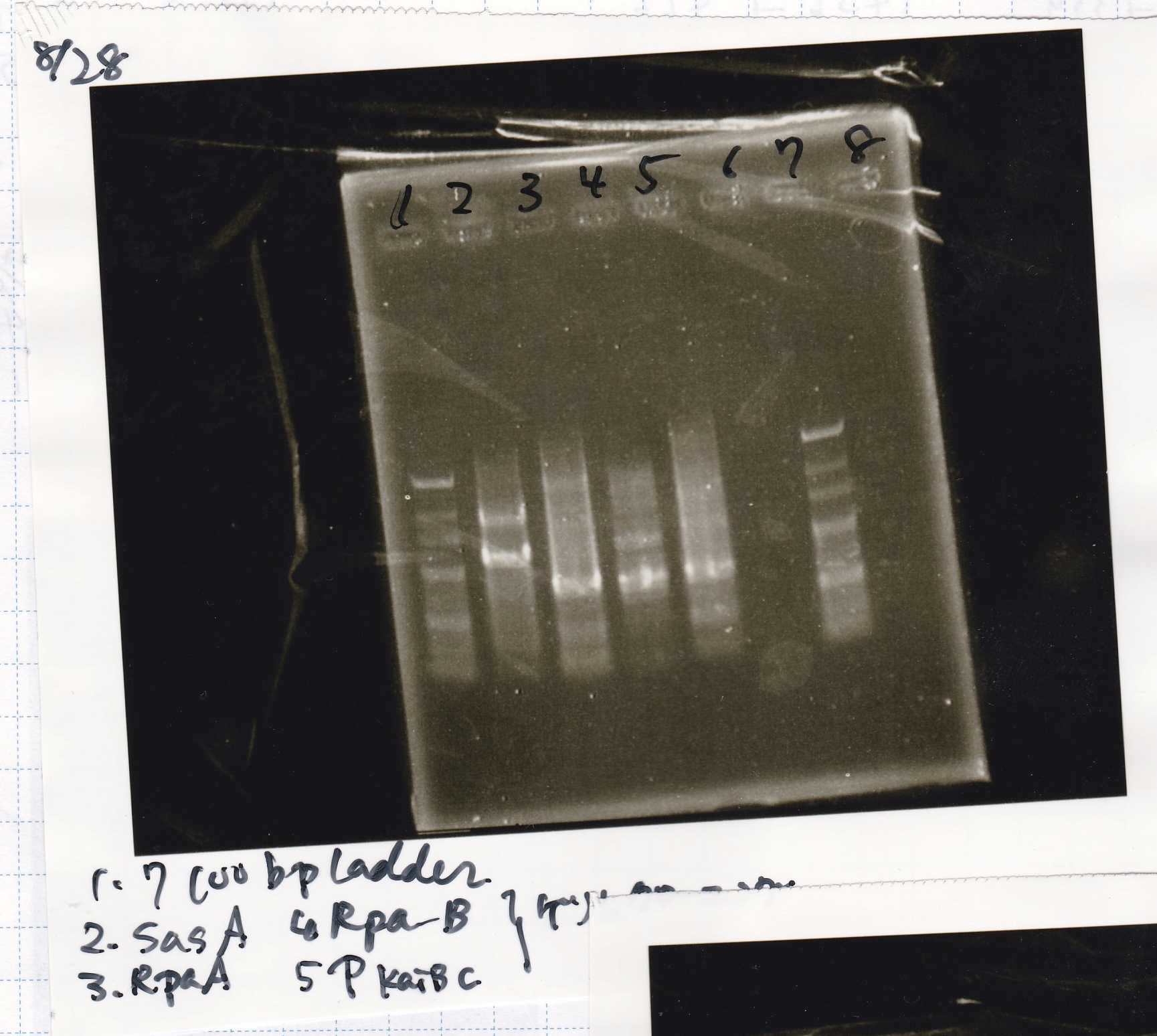
Aug 28
Electrophoresis
No name
| Lane | Sample
|
| 1 | 100bp
|
| 2 | SasA
|
| 3 | RpaA
|
| 4 | RpaB
|
| 5 | PkaiBC
|
| 6 | PCR NC
|
| 7 | 100bp
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 8/27 Plux | 173.8 | 1.95 | 1.45
|
Ligation
No name
| state | Vector | Inserter
|
| experiment | 8/17 DT-1 | 3.0 | 8.0
|
- Samples were evaporeted used evaporator into about 3 µL.
| sample | MilliQ | Ligation High | total
|
| 3 | 4 | 3.5 | 10.5
|
- incubate one hour at 16 °C
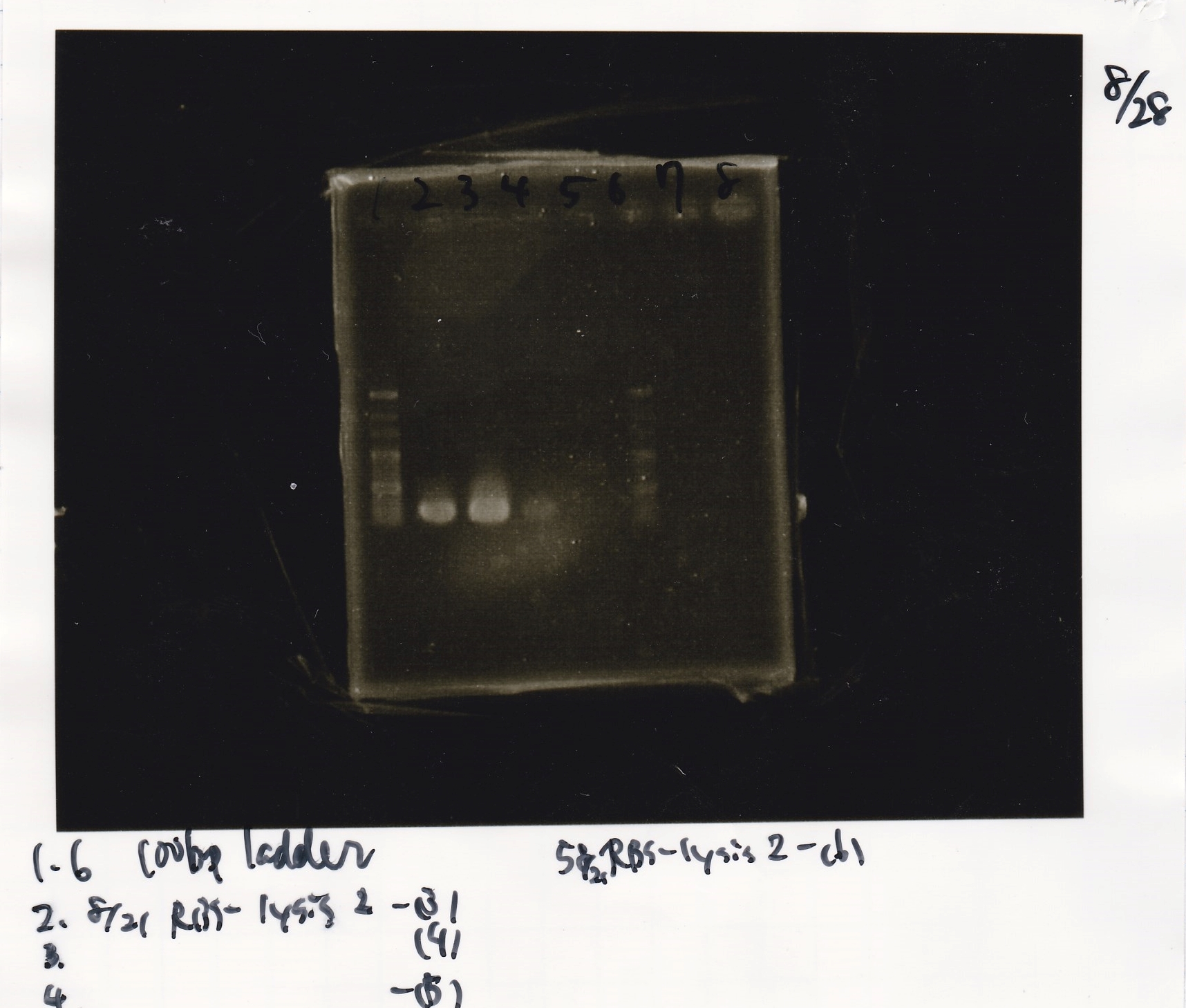
No name
| Lane | DNA
|
| 1 | 100bpladder
|
| 3 | SasA(8/27 Genome PCR production)
|
| 5 | RpaA(8/27 Genome PCR production)
|
| 7 | RpaB(8/27 Genome PCR production)
|
| 9 | PkaiBC(8/27 Genome PCR production)
|


| Name | concentration[µg/mL] | 260/280 | 260/230
|
| RpaA | 15.4 | 1.88 | 0.03
|
| SasA | 21.6 | 1.66 | 0.79
|
| PkaiBC | 16.3 | 1.79 | 0.68
|
| RpaB | 26.7 | 1.77 | 0.82
|
Colony PCR
No name
| Sample | base pair
|
| 8/21 RBS-lysis2-(3) | 772
|
| 8/21 RBS-lysis2-(4) | 772
|
| 8/21 RBS-lysis2-(5) | 772
|
| 8/21 RBS-lysis2-(6) | 772
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 48s | 30cycles
|
Restriction Enzyme Digestion
No name
| | 8/22 RBS-lysis1-(1) | EcoRI | SpeI | BufferB | BSA | MilliQ | total
|
| 2 cuts | 9µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 16.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
| | 8/17 RBS-GFP-DT-(2) | XbaI | PstI | BufferD | BSA | MilliQ | total
|
| 2 cuts | 4µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 21.7µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
| | 8/20 Pcon-RBS-GFP-DT-(1) | EcoRI | SpeI | BufferB | BSA | MilliQ | total
|
| 2 cuts | 3µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 22.7µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 0.1µL | 8.6µL | 10µL
|
| | 8/20 Pcon-RBS-luxR-DT-(1) | EcoRI | XbaI | BufferD | BSA | MilliQ | total
|
| 2 cuts | 2µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 23.7µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 0.1µL | 8.6µL | 10µL
|
No name
| | 8/28 Plux | SpeI | PstI | BufferD | BSA | MilliQ | total
|
| 2 cuts | 6µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 19.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
| | 8/20 RBS-tetR-DT-(1) | XbaI | PstI | BufferD | BSA | MilliQ | total
|
| 2 cuts | 5µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 20.7µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
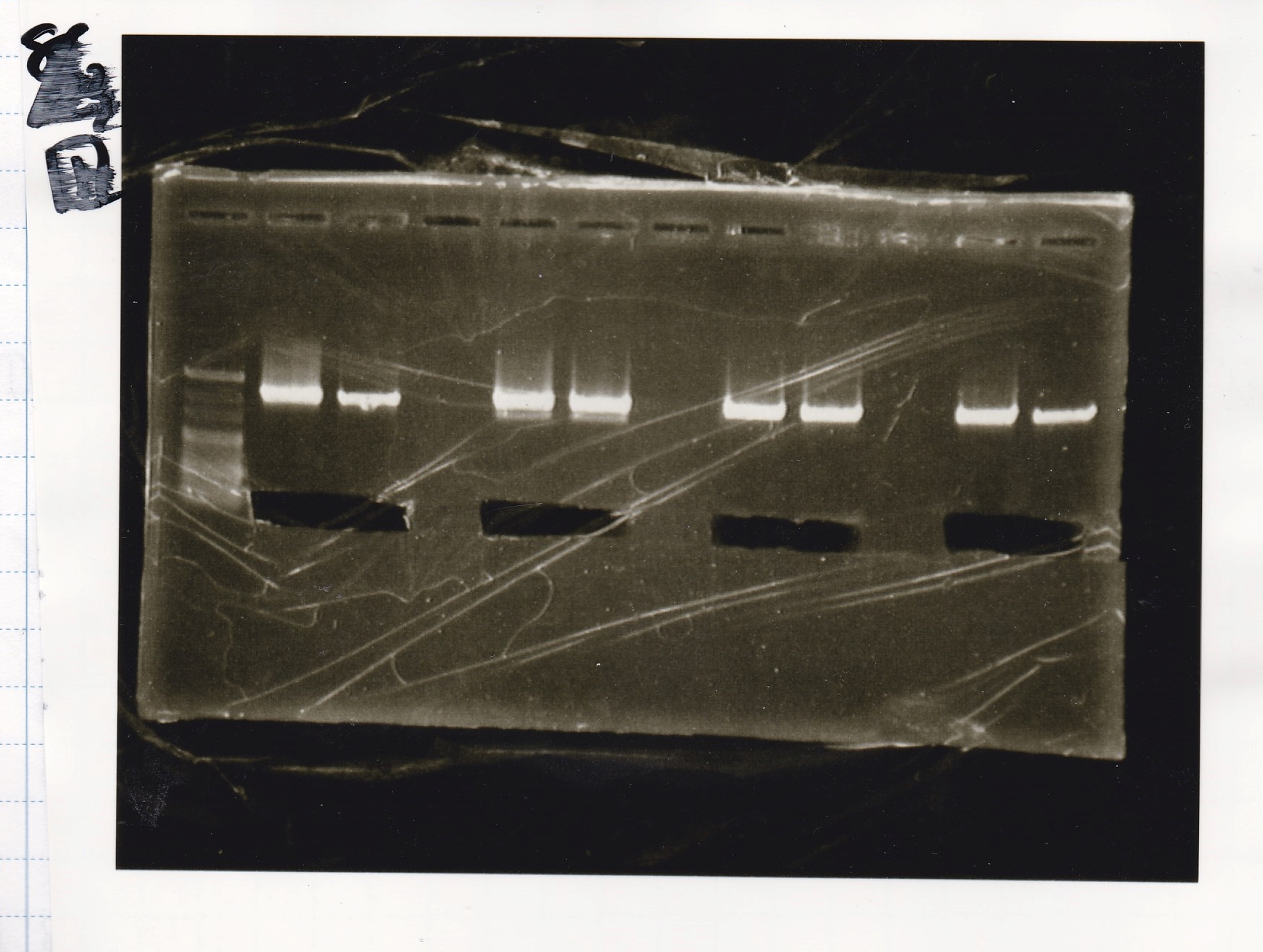
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 8/22 RBS-lysis1-(1) | EcoRI | SpeI
|
| 2 | 8/22 RBS-lysis1-(1) | -- | --
|
| 3 | 8/17 RBS-GFP-DT-(2) | XbaI | PstI
|
| 4 | 8/17 RBS-GFP-DT-(2) | -- | --
|
| 5 | 8/20 Pcon-RBS-GFP-DT-(1) | EcoRI | SpeI
|
| 6 | 8/20 Pcon-RBS-GFP-DT-(1) | -- | --
|
| 7 | 100bp ladder | -- | --
|
| 8 | 8/20 Pcon-RBS-luxR-DT-(1) | EcoRI | XbaI
|
| 9 | 8/20 Pcon-RBS-luxR-DT-(1) | -- | --
|
| 10 | 8/28 Plux | SpeI | PstI
|
| 11 | 8/28 Plux | -- | --
|
| 12 | 8/20 RBS-tetR-DT | XbaI | PstI
|
| 13 | 8/20 RBS-tetR-DT | -- | --
|
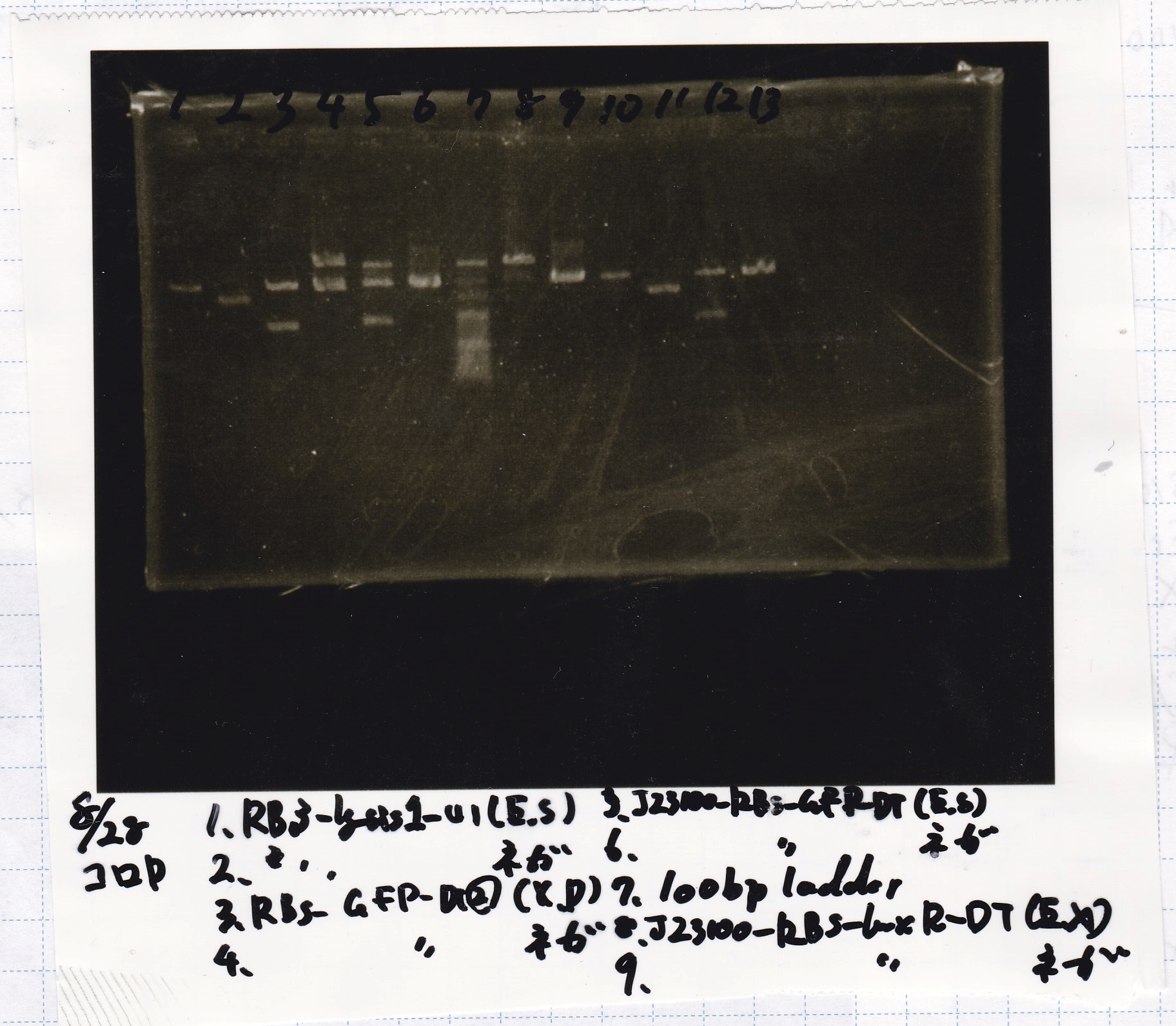
No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | 8/20 RBS-lysis1-(1) | EcoRI & SpeI
|
| 3
|
| 4 | -- | --
|
| 5 | 8/28 RBS-GFP-DT-(2) | XbaI & PstI
|
| 6
|
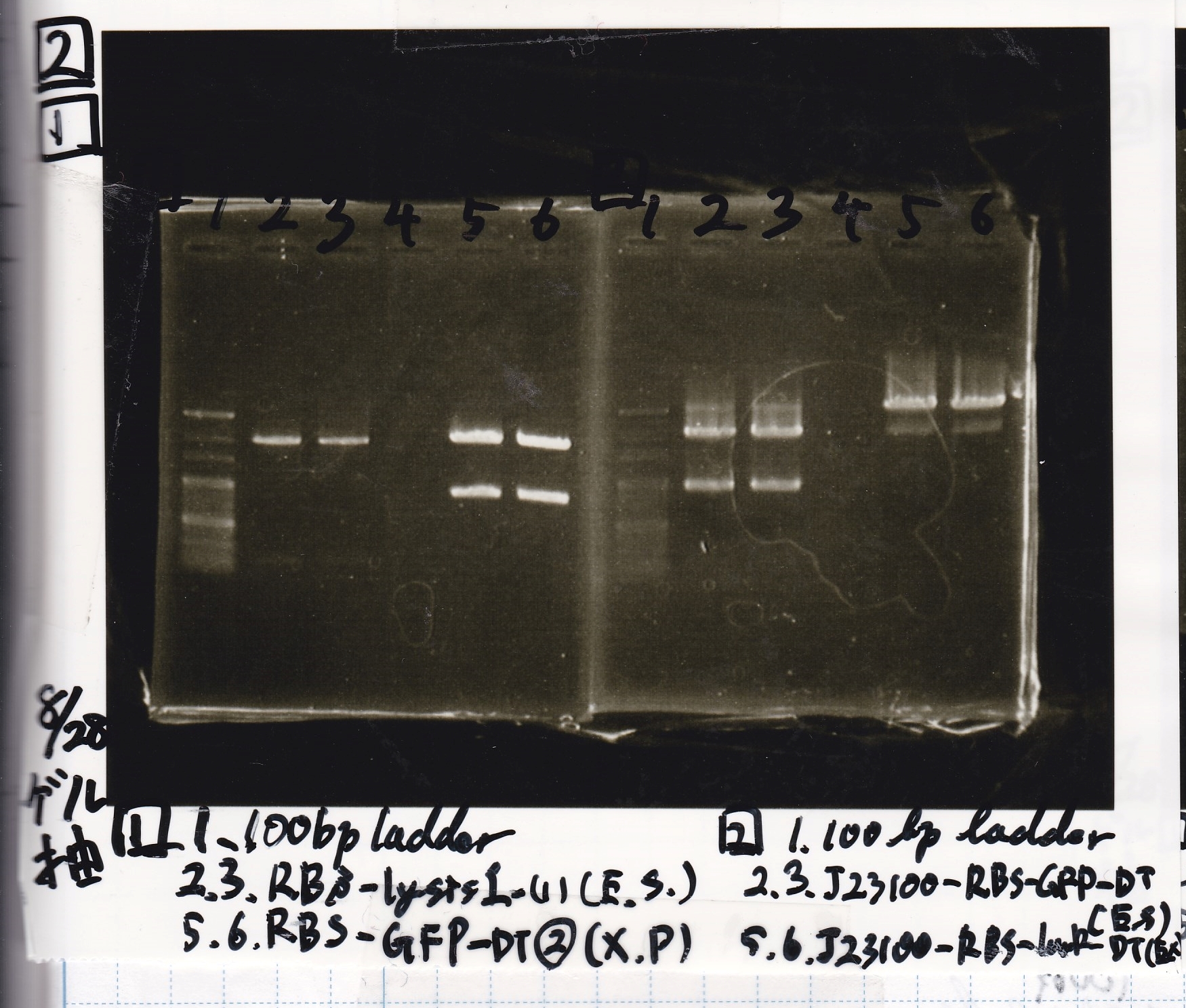
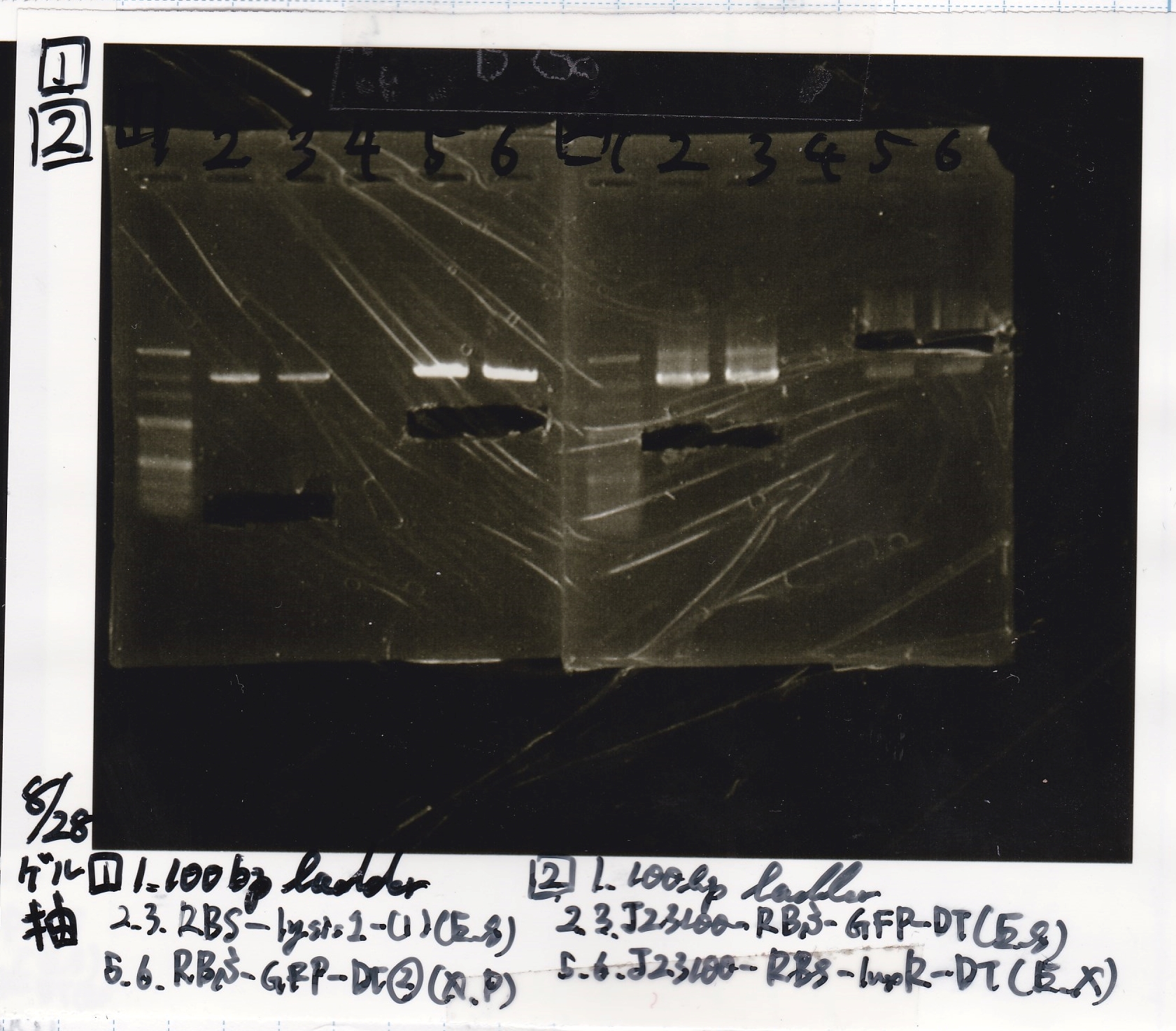
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | 8/28 Plux | SpeI & PstI
|
| 3
|
| 4 | -- | --
|
| 5 | 8/20 RBS-tetR-DT-(1) | XbaI & PstI
|
| 6
|
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-RBS-GFP-DT (EcoRI&SpeI) | 7.1 | 1.88 | 0.07
|
| Pcon-RBS-luxR-DT (EcoRI&XbaI) | 15.3 | 1.93 | 0.70
|
| Plux (SpeI&PstI) | 4.5 | 1.79 | 0.31
|
| RBS-GFP-DT (XbaI&PstI) | 6.0 | 2.16 | 0.03
|
| RBS-tetR-DT (XbaI&PstI) | 7.6 | 1.91 | 0.38
|
Colony PCR
No name
| Sample | base pair
|
| 8/27 pT181 attenuator-DT-1 | 738
|
| 8/27 pT181 attenuator-DT-2 | 738
|
| 8/27 pT181 attenuator-DT-3 | 738
|
| 8/27 TetR-aptamer 12_1R-DT-1 | 521
|
| 8/27 TetR-aptamer 12_1R-DT-2 | 521
|
| 8/27 pT181 antisense-DT-1 | 542
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
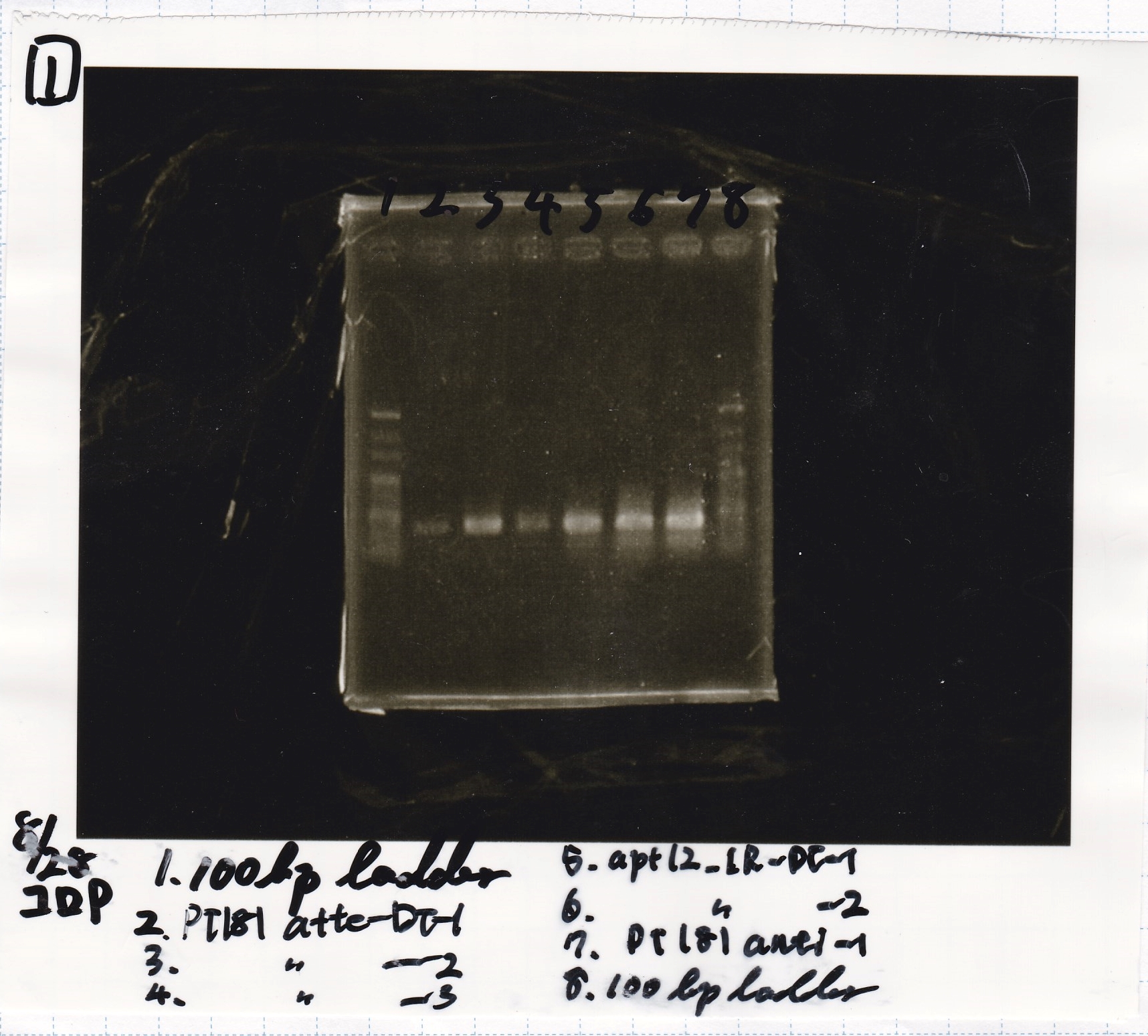
| Sample | base pair
|
| 8/27 Pλ-luxI-1 | --
|
| 8/27 Pλ-luxI | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30min | 3cycles
|

Colony PCR
No name
| Sample | base pair
|
| 8/21 RBS-lysis2-9 | 772
|
| 8/21 RBS-lysis2-10 | 772
|
| 8/21 RBS-lysis2-11 | 772
|
| 8/21 RBS-lysis2-12 | 772
|
| 8/21 RBS-lysis2-13 | 772
|
| 8/21 RBS-lysis2-14 | 772
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 48s | 30cycles
|
Liquid Culture
No name
| Sample | medium
|
| 8/16 Master Plate(CP)-6 DT | Plusgrow medium(+CP)
|
| J23100 Master Plate-1 | Plusgrow medium(+Amp)
|
| Plac Master Plate-1 | Plusgrow medium(+CP)
|
Ligation
No name
| state | Vector | Inserter
|
| experiment | 8/27 DT (EcoRI & XbaI) | 2.9 | 8/28 RBS-lysis1 (EcoRI & SpeI) | 9.5
|
| experiment | 8/28 Plux (SpeI & PstI) | 2.3 | 8/28 RBS-GFP-DT (XbaI & PstI) | 18
|
| experiment | 8/28 Pcon-RBS-luxR-DT (EcoRI & XbaI) | 3.3 | 8/28 Pcon^RBS-GFP-DT-(1) (EcoRI & SpeI) | 28
|
| experiment | 8/27 DT (EcoRI & XbaI) | 2.7 | 8/24 Spinach (EcoRI & SpeI) | 2.4
|
- Samples were evaporeted used evaporator into about 1 µL.
| sample | MilliQ | Ligation High | total
|
| 1 | 3 | 4 | 9
|
- incubate overnight at 4 °C
Liquid Culture
No name
| Sample | medium
|
| 8/26 Pλ-RBS-luxI-DT-1 | Plusgrow medium (+Amp)
|
| 8/26 Pλ-RBS-luxI-DT-2 | Plusgrow medium (+Amp)
|
Aug 29
Colony PCR
No name
| Sample | base pair
|
| 8/27 Pcon-pT181 antisense-(1) | 542
|
| 8/26 pT181 attenuator-DT-(1) | 738
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | 8/27 Pcon-pT181antisense-1
|
| 3 | 8/26 pT181attenuator-DT-1
|
| 4 | 100bp ladder
|

Electrophoresis
No name
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | 8/21 RBS-lysis2-9(Colony PCR prodution)
|
| 3 | 8/21 RBS-lysis2-8(Colony PCR prodution)
|
| 4 | 8/21 RBS-lysis2-7(Colony PCR prodution)
|
| 5 | 8/21 RBS-lysis2-10(Colony PCR prodution)
|
| 6 | 8/21 RBS-lysis2-11(Colony PCR prodution)
|
| 7 | 8/21 RBS-lysis2-12(Colony PCR prodution)
|
| 8 | 8/21 RBS-lysis2-13(Colony PCR prodution)
|
| 9 | 8/21 RBS-lysis2-14(Colony PCR prodution)
|
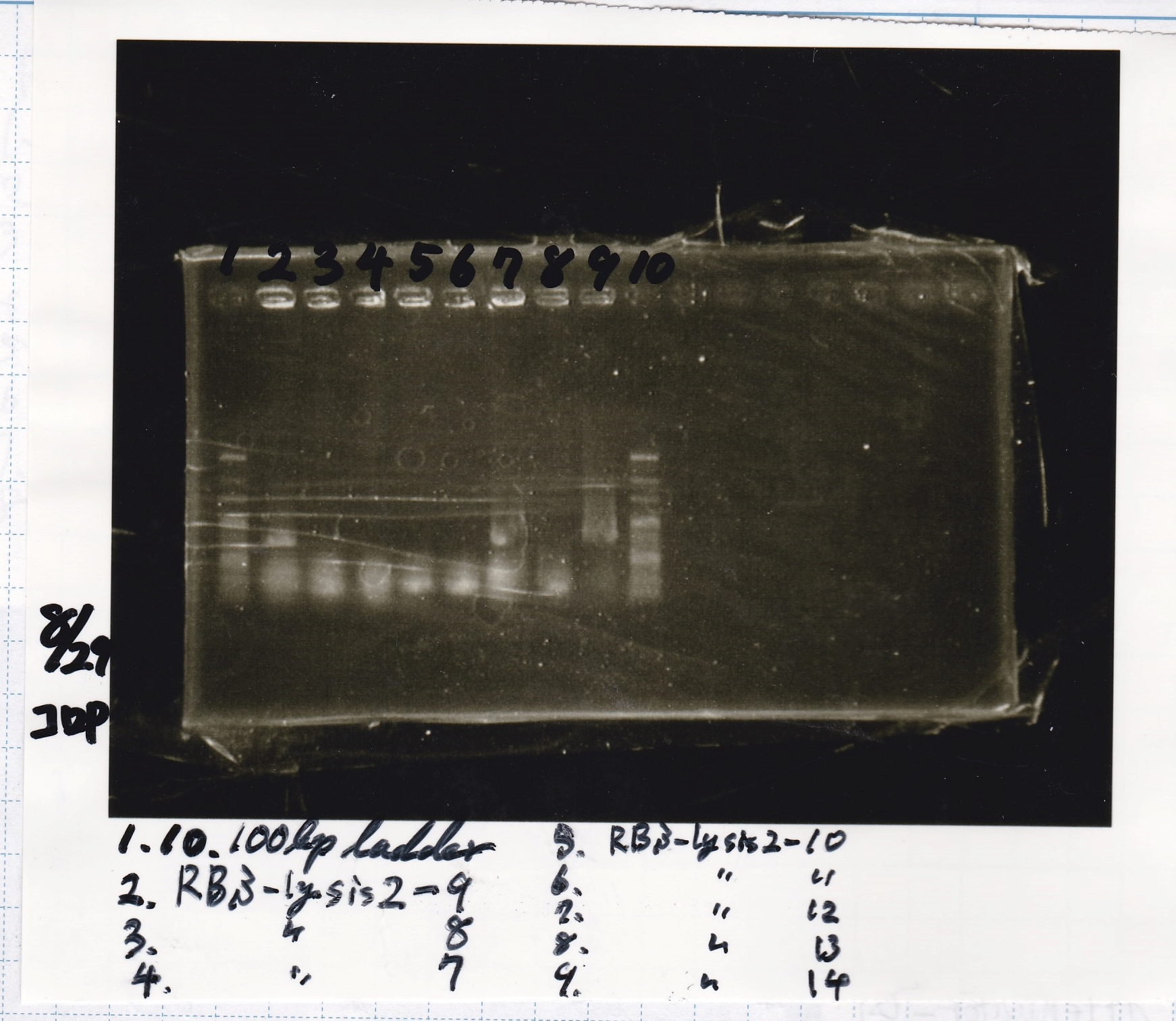
Miniprep
No Name
| DNA | concentration [µg/mL] | 260/280 | 260/230
|
| Plac | 153.7 | 2.03 | 2.10
|
| Pλ-luxⅠ-(1) | 76.5 | 2.07 | 2.06
|
| Pλ-luxⅠ-(2) | 86.4 | 2.08 | 2.10
|
| DT | 166.2 | 1.65 | 2.12
|
| J23100 | 190.1 | 1.68 | 2.34
|
Liquid Culture
No name
| Sample | medium
|
| 8/21 RBS-Lysis2-9 | Plusgrow medium(+Amp)
|
| 8/21 RBS-Lysis2-12 | Plusgrow medium(+Amp)
|
| 8/21 RBS-Lysis2-14 | Plusgrow medium(+Amp)
|
Restriction Enzyme Digestion
No name
| | 8/22 psB1C3-(2) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2 cuts | 8µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 17.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 7.9µL | 10µL
|
| 2 cuts | 8µL | 0µL | 0µL | 0.5µL | 0.5µL | 0µL | 3µL | 0.3µL | 17.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
| | 8/21 tRMA-spinach(1) | XbaI | SpeI | BufferB | BSA | MilliQ | total
|
| 2 cuts | 4µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 21.7µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
| | 8/21 tetR aptamer12-1R(1) | EcoRI | SpeI | BufferB | BSA | MilliQ | total
|
| 2 cuts | 3µL | 0.5µL | 0.5µL | 3µL | 0.3µL | 22.7µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 0.1µL | 8.5µL | 10µL
|
| | 8/21 PT181 attenuator-(2) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2 cuts | 4µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 21.7µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.4µL | 10µL
|
| 2 cuts | 4µL | 0µL | 0µL | 0.5µL | 0.5µL | 0µL | 3µL | 0.3µL | 21.7µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
| | 8/21 PT181 antisense-(2) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2 cuts | 10µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 15.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 7.9µL | 10µL
|
| 2 cuts | 10µL | 0µL | 0µL | 0.5µL | 0.5µL | 0µL | 3µL | 0.3µL | 15.7µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
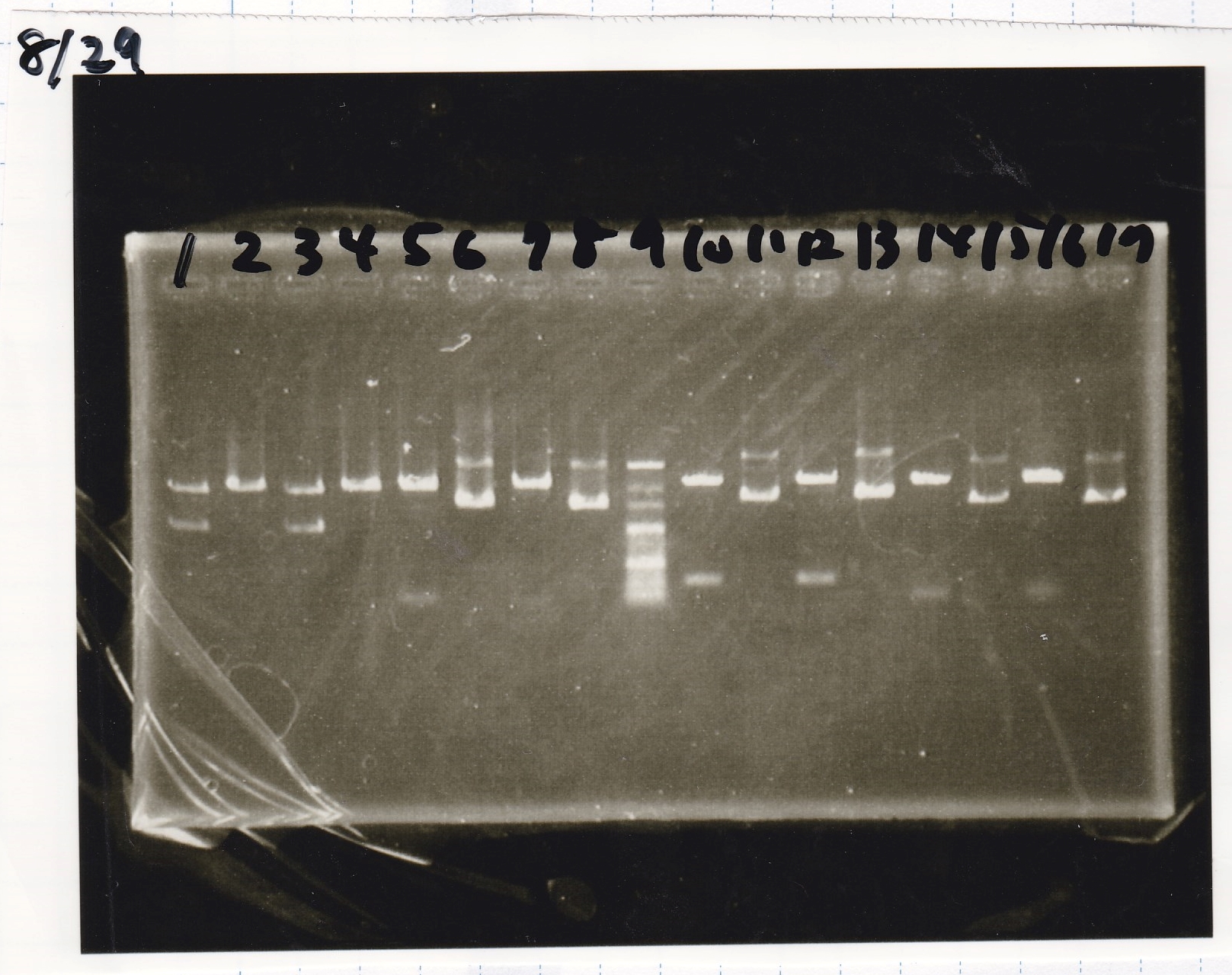
Transformation
No Name
| Name | Sample | Competent Cells | Total | Plate
|
| 8/27 pT181 attenuator-DT | 2µL | 20µL | 22µL | CP
|
| 8/27 RBS-lysis3-DT | 2µL | 20µL | 22µL | CP
|
| 8/28 RBS-lysis3-DT | 2µL | 20µL | 22µL | CP
|
| 8/28 Pcon-RBS-GFP-DT-Pcon-RBS-LuxR-DT | 2µL | 20µL | 22µL | Amp
|
| 8/28 RBS-lysis1-DT | 2µL | 20µL | 22µL | CP
|
| 8/28 Plux-RBS-GFP-DT | 2µL | 20µL | 22µL | CP
|
| 8/28 Spinach-DT | 2µL | 20µL | 22µL | CP
|
No name
| Lane | DNA | Enzyme1 | Enzyme2
|
| 100bp ladder | -- | --
|
| 9 | 8/29 pT181attenuator(2) | EcoRI | SpeI
|
| 10 | 8/29 pT181attenuator(2) | EcoRI | SpeI
|
| 11 | 8/29 pT181attenuator(2) | XbaI | PstI
|
| 12 | 8/29 pT181attenuator(2) | XbaI | PstI
|
| 13 | pT181antisense(2) | EcoRI | SpeI
|
| 14 | pT181antisense(2) | EcoRI | SpeI
|
| 15 | pT181antisense(2) | XbaI | PstI
|
| 16 | pT181antisense(2) | XbaI | PstI
|
| 100bp ladder | -- | --
|

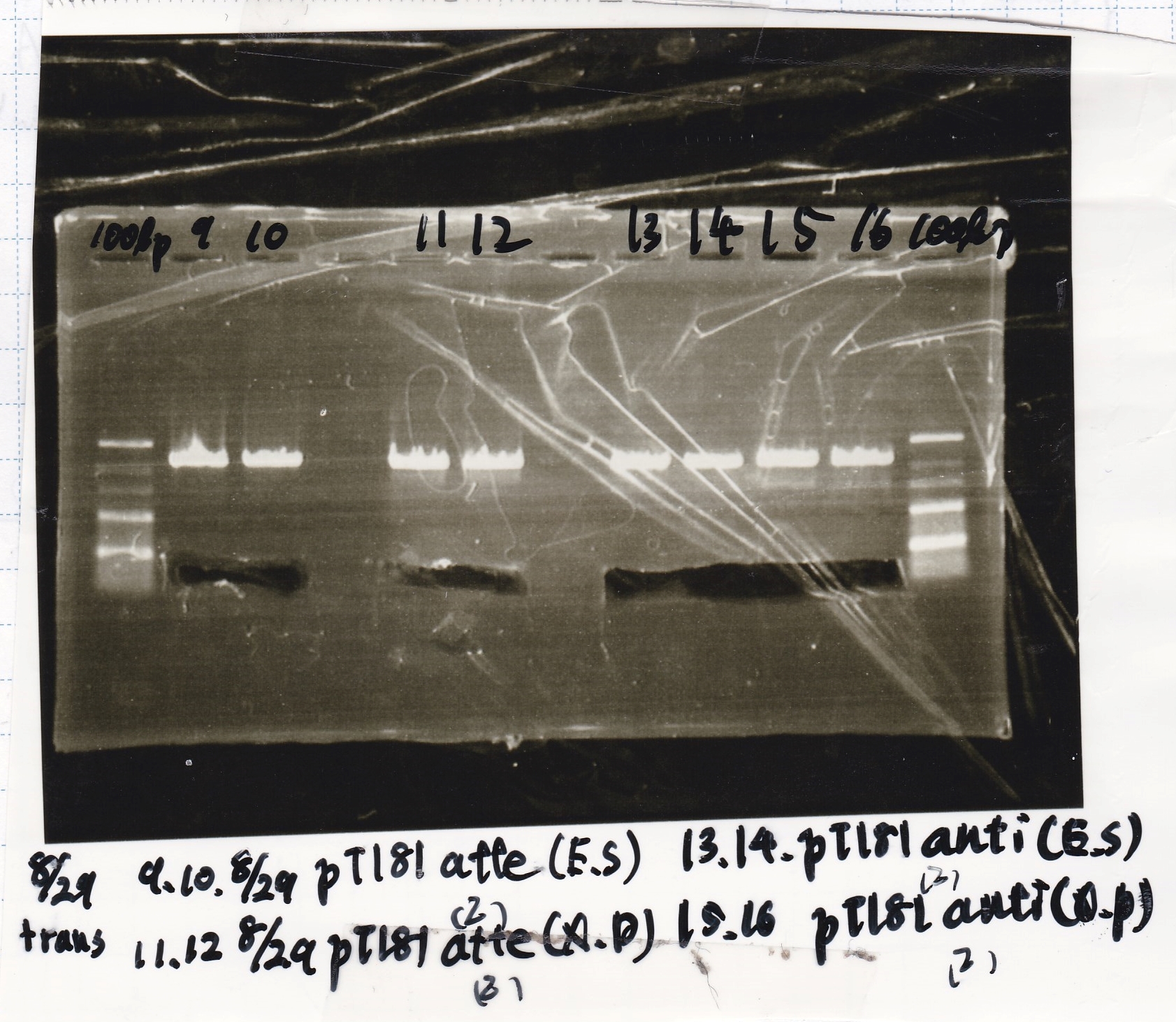
| Lane | Sample | Enzyme1 | Enzyme2
|
| 100bp ladder | -- | --
|
| 1 | 8/29 pSB1C3 | EcoRI | SpeI
|
| 2 | 8/29 pSB1C3 | EcoRI | SpeI
|
| 3 | 8/29 pSB1C3 | XbaI | PstI
|
| 4 | 8/29 pSB1C3 | XbaI | PstI
|
| 5 | 8/29 spinach | EcoRI | SpeI
|
| 6 | 8/29 spinach | EcoRI | SpeI
|
| 7 | 8/29 tetRaptarmer12-1R(1) | EcoRI | SpeI
|
| 8 | 8/29 tetRaptarmer12-1R(1) | EcoRI | SpeI
|
| 100bp ladder | -- | --
|
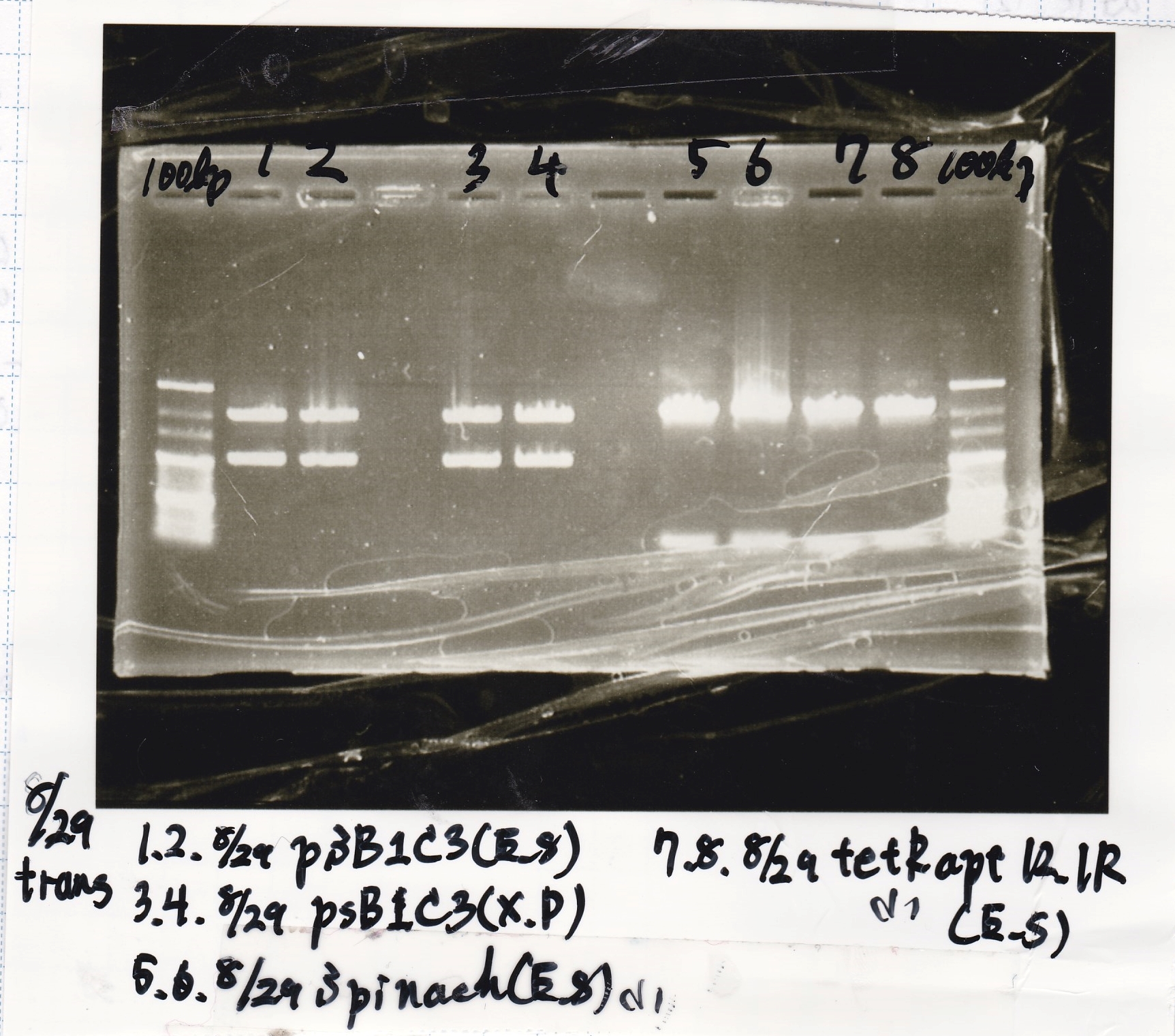

Aug 30
No Name
| Name | quantity | concentration[µg/mL] | 260/280 | 260/230
|
| pT181 attenuator(2) (EcoRI & SpeI) | 305.3 | 2.2 | 1.66 | 0.36
|
| pT181 attenuator(2) (XbaI & PstI) | 217.7 | 3.9 | 1.73 | 0.04
|
| pT181 antisense(2) (EcoRI & SpeI) | 347.7 | 3.1 | 1.88 | 0.31
|
| pT181 antisense(2) (XbaI & PstI) | 376.5 | 5.9 | 0.49 | 2.36
|
| pSB1C3 (EcoRI & SpeI) | 436.6 | 30.7 | 1.03 | 1.28
|
| pSB1C3 (XbaI & PstI) | 457.4 | 7.9 | 1.65 | 0.42
|
| Spinach (EcoRI & SpeI) | 490.5 | 3.0 | 1.87 | 0.26
|
| tetR aptamer 12_1R (EcoRI & SpeI) | 451.7 | -12.8 | 5.56 | -0.04
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-lysis2 9 | 264.6 | 1.97 | 2.21
|
| RBS-lysis2 12 | 220.0 | 1.89 | 2.46
|
| RBS-lysis2 14 | 195.3 | 1.88 | 1.53
|
Colony PCR
No name
| Sample | base pair
|
| 8/29 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT(8/20 LB Amp) (1)~(4) | 2143
|
| 8/29 Plux-RBS-GFP-DT(8/27 LB-CP) (1)~(2) | 1227
|
| 8/29 Pbad/araC-RBS-RFP{BBa_I13516} (8/21 LB-CP) (1)~(2) | 2256
|
| 8/29 RBS-lysis3-DT-(1) | 1210
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 2min | 30cycle
|

Ethanol Precipitation
No name
| DNA
|
| pT181 attenuator-(2) (EcoRI & SpeI)
|
| pT181 attenuator-(2) (XbaI & PstI)
|
| pT181 antisense-(2) (EcoRI & SpeI)
|
| pT181 antisense-(2) (XbaI & PstI)
|
| pSB1C3 (EcoRI & SpeI)
|
| pSB1C3 (XbaI & PstI)
|
| Spinach (EcoRI & SpeI)
|
| tetR aptamer 12_1R (EcoRI & SpeI)
|
Restriction Enzyme Digestion
No name
| | 8/30 RBS-lysis2 (9) | EcoRI | SpeI | 10x bufferB | 100x BSA | MilliQ | total
|
| 2 cuts | 7.5 µL | 0.5 µL | 0.5 µL | 3 µL | 0.3 µL | 18.2 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 1 µL | 0.1 µL | 8.4 µL | 10 µL
|
| | 8/22 pSB1C3-(2) | EcoRI | SpeI | 10x Buffer | 100x BSA | MilliQ | total
|
| 2 cuts | 8 µL | 0.5 µL | 0.5 µL | 3 µL | 0.3 µL | 17.7 µL | 30 µL
|
| NC | 1 µL | 0 µL | 0 µL | 1 µL | 0.1 µL | 7.9 µL | 10 µL
|
| | 8/22 pSB1C3-(2) | XbaI | PstI | BufferD | BSA | MilliQ | total
|
| 2 cuts | 8 µL | 0.5 µL | 0.5 µL | 3 µL | 0.3 µL | 17.7 µL | 30 µL
|
| NC | 1 µL | 0 µL | 0 µL | 1 µL | 0.1 µL | 7.9 µL | 10 µL
|
| | 8/21 Spinach (1) | EcoRI | SpeI | 10x BufferB | 100x BSA | MilliQ | total
|
| 2 cuts | 4 µL | 0.5 µL | 0.5 µL | 3 µL | 0.3 µL | 21.9 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 1 µL | 0.1 µL | 8.4 µL | 10 µL
|
| | 8/21 tetR aptamer 12_1R (1) | EcoRI | SpeI | 10xBufferB | 100xBSA | MilliQ | total
|
| 2 cuts | 3 µL | 0.5 µL | 0.5 µL | 3 µL | 0.3 µL | 22.7 µL | 30 µL
|
| NC | 0.4 µL | 0 µL | 0 µL | 1 µL | 0.1 µL | 8.4 µL | 10 µL
|
| | 8/21 pT181 attenuator-(2) | EcoRI | SpeI | XbaI | PstI | 10x BufferB | 10x BufferD | 100x BSA | MilliQ | total
|
| 2 cuts | 4 µL | 0.5 µL | 0.5 µL | 0 µL | 0 µL | 3 µL | 0 µL | 0.3 µL | 21.7 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 0 µL | 0 µL | 1 µL | 0 µL | 0.1 µL | 8.4 µL | 10 µL
|
| 2 cuts | 4 µL | 0 µL | 0 µL | 0.5 µL | 0.5 µL | 0 µL | 3 µL | 0.3 µL | 21.7 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 0 µL | 0 µL | 0 µL | 1 µL | 0.1 µL | 8.4 µL | 10 µL
|
| | 8/21 pT181 antisense-(2) | EcoRI | SpeI | XbaI | PstI | 10x BufferB | 10x BufferD | 100x BSA | MilliQ | total
|
| 2 cuts | 9 µL | 0.5 µL | 0.5 µL | 0 µL | 0 µL | 3 µL | 0 µL | 0.3 µL | 16.7 µL | 30 µL
|
| NC | 1 µL | 0 µL | 0 µL | 0 µL | 0 µL | 1 µL | 0 µL | 0.1 µL | 7.9 µL | 10 µL
|
| 2 cuts | 9 µL | 0 µL | 0 µL | 0.5 µL | 0.5 µL | 0 µL | 3 µL | 0.3 µL | 16.7 µL | 30 µL
|
| NC | 1 µL | 0 µL | 0 µL | 0 µL | 0 µL | 0 µL | 1 µL | 0.1 µL | 7.9 µL | 10 µL
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | RBS-lysis2-(9) | EcoRI | SpeI
|
| 3 | RBS-lysis2-(9) | -- | --
|
| 4 | pSB1C3-(2) | EcoRI | SpeI
|
| 5 | pSB1C3-(2) | -- | --
|
| 6 | pSB1C3-(2) | XbaI | PstI
|
| 7 | pSB1C3-(2) | -- | --
|
| 8 | tRNA-Spinach-(1) | EcoRI | SpeI
|
| 9 | tRNA-Spinach-(1) | -- | --
|
| 10 | tetR aptamer12_1R | EcoRI | SpeI
|
| 11 | tetR aptamer12_1R | -- | --
|
| 12 | pT181 attenuator | EcoRI | SpeI
|
| 13 | pT181 attenuator | EcoRI | SpeI
|
| 14 | 100bp ladder | -- | --
|
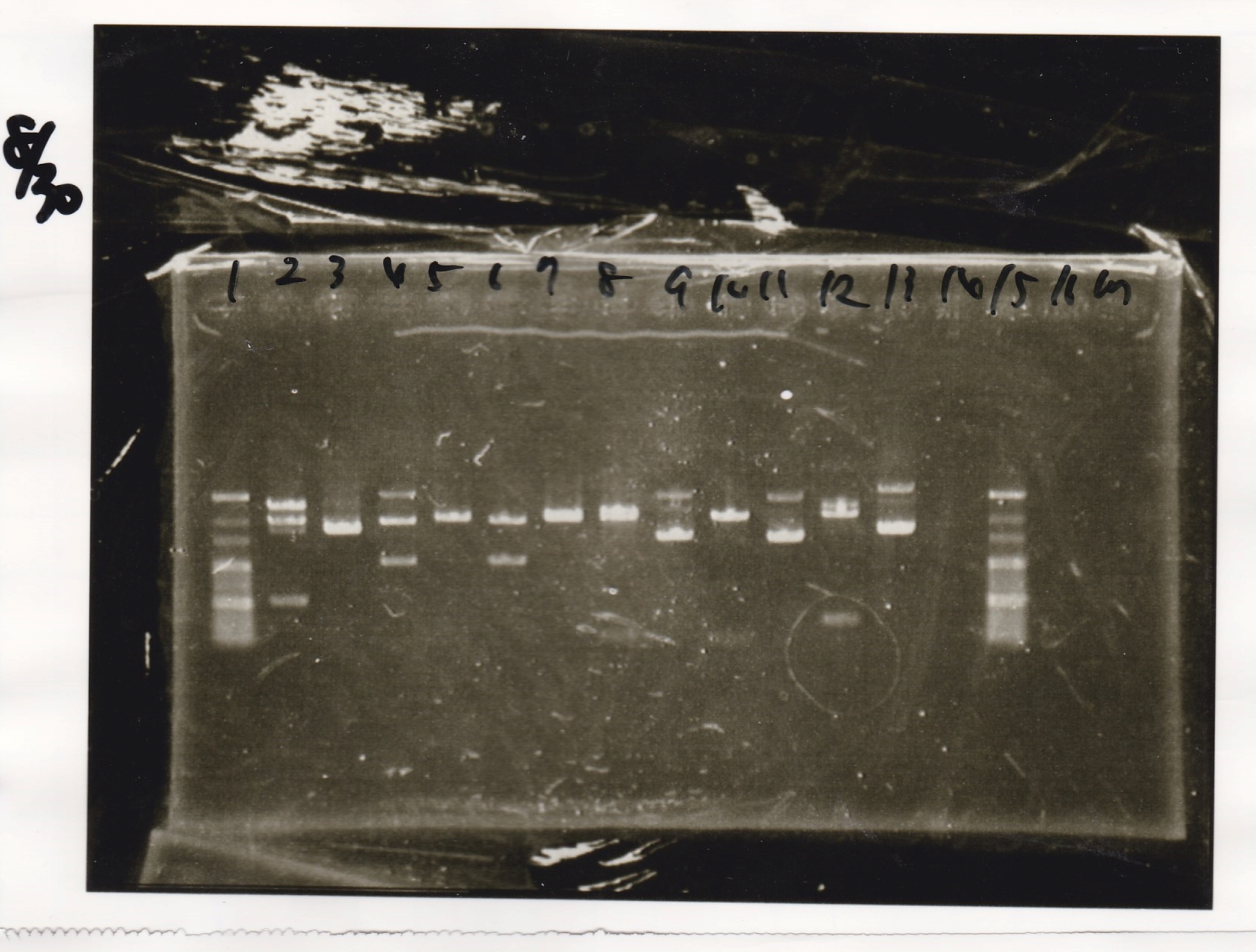
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | pT181 antisense | EcoRI | SpeI
|
| 3 | pT181 antisense | -- | --
|
| 4 | pT181 antisense | XbaI | PstI
|
| 5 | pT181 antisense | -- | --
|
| 6 | pT181 attenuator | XbaI | PstI
|
| 7 | pT181 attenuator | -- | --
|
| 8 | 100bp ladder | -- | --
|
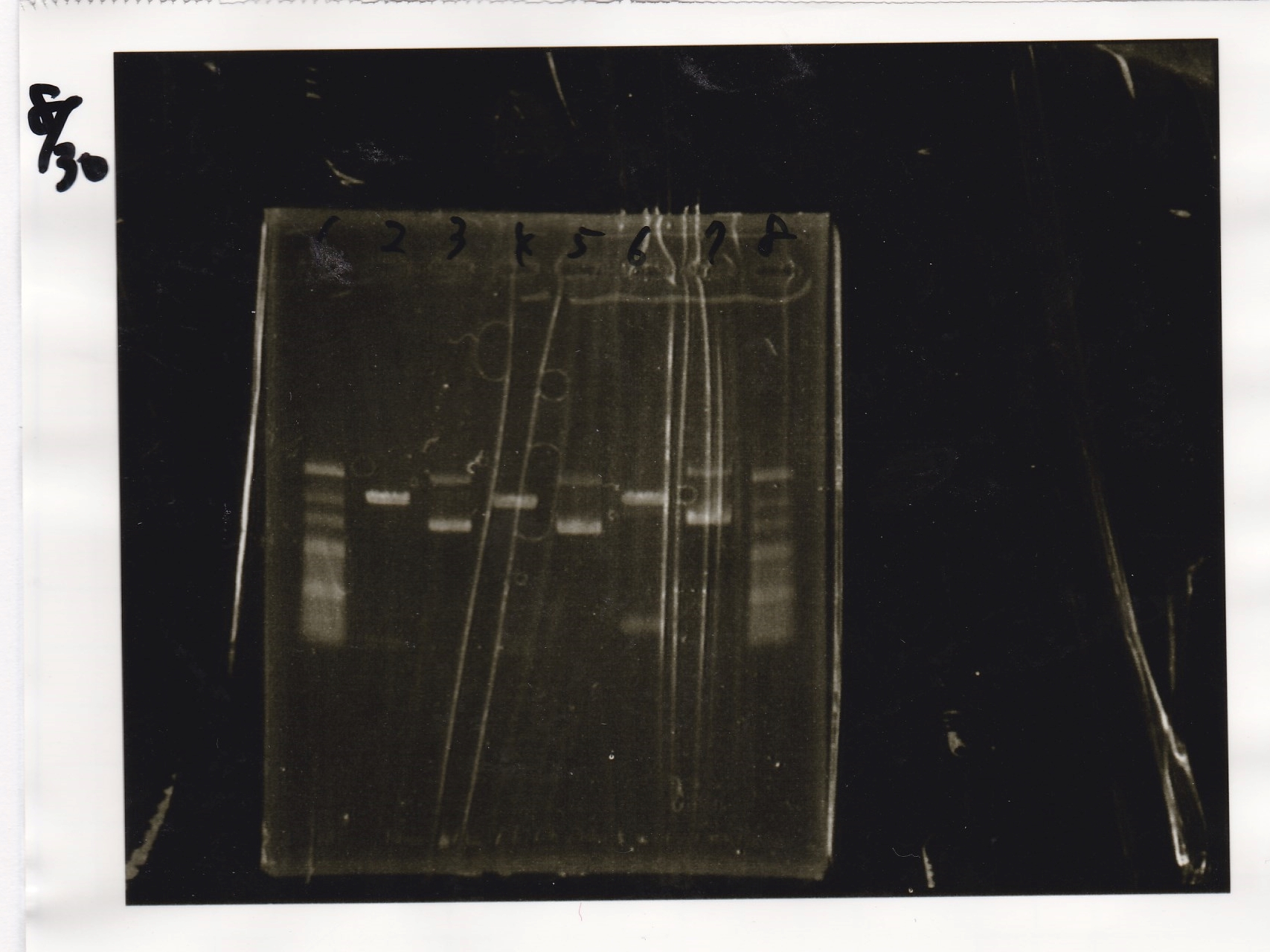
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | 8/29 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(1)(Colony PCR product)
|
| 3 | 8/29 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(2)(Colony PCR product)
|
| 4 | 8/29 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(3)(Colony PCR product)
|
| 5 | 8/29 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(4)(Colony PCR product)
|
| 6 | Pbad/araC-RBS-RFP-DT-(1)(Colony PCR product)
|
| 7 | Pbad-araC-RBS-RFP-DT-(2)(Colony PCR product)
|

Liquid Culture
No name
| Sample | medium
|
| 8/29 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(1) | Plusgrow medium(+Amp)
|
| 8/29 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(2) | Plusgrow medium(+Amp)
|
| 8/29 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(3) | Plusgrow medium(+Amp)
|
| 8/29 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(4) | Plusgrow medium(+Amp)
|
| 8/29 Plux-RBS-GFP-DT-(1) | Plusgrow medium(+CP)
|
| 8/29 Plux-RBS-GFP-DT-(2) | Plusgrow medium(+CP)
|
| 8/29 Pbad/araC-RBS-RFP-DT-(1) | Plusgrow medium(+CP)
|
| 8/29 Pbad-araC-RBS-RFP-DT-(2) | Plusgrow medium(+CP)
|
| RBS-lysis3-DT-(1) | Plusgrow medium(+CP)
|
Aug 31
No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | -- | --
|
| 3 | RBS-lysis2-(9) | EcoRI & SpeI
|
| 4
|
| 5 | -- | --
|
| 6 | pSB1C3(1) | EcoRI & SpeI
|
| 7
|
| 8 | -- | --
|
| 9 | pSB1C3(1) | XbaI & PstI
|
| 10
|

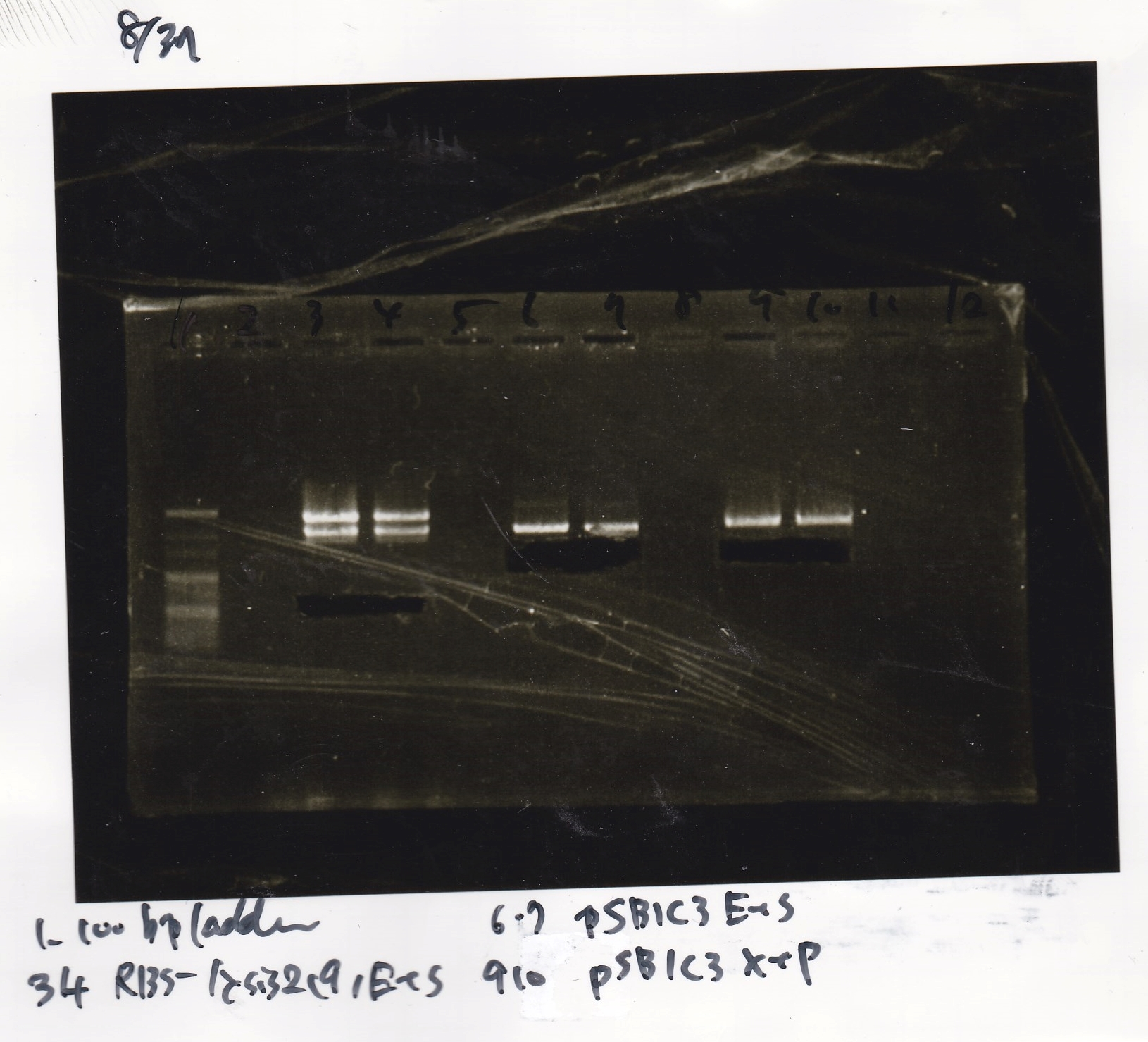
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | tRNA-Spinach | EcoRI & SpeI
|
| 3
|
| 4 | -- | --
|
| 5 | pT181 attenuator-(2) | EcoRI & SpeI
|
| 6
|
| 7 | pT181 attenuator-(2) | XbaI & PstI
|
| 8
|
| 9 | -- | --
|
| 10 | pT181 antisense-(2) | EcoRI & SpeI
|
| 11
|


| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | pT181 antisense | XbaRI & PstI
|
| 3
|
| 4 | -- | --
|
| 5 | tetR aptamer 12_1R-(1) | EcoRI & SpeI
|
| 6
|
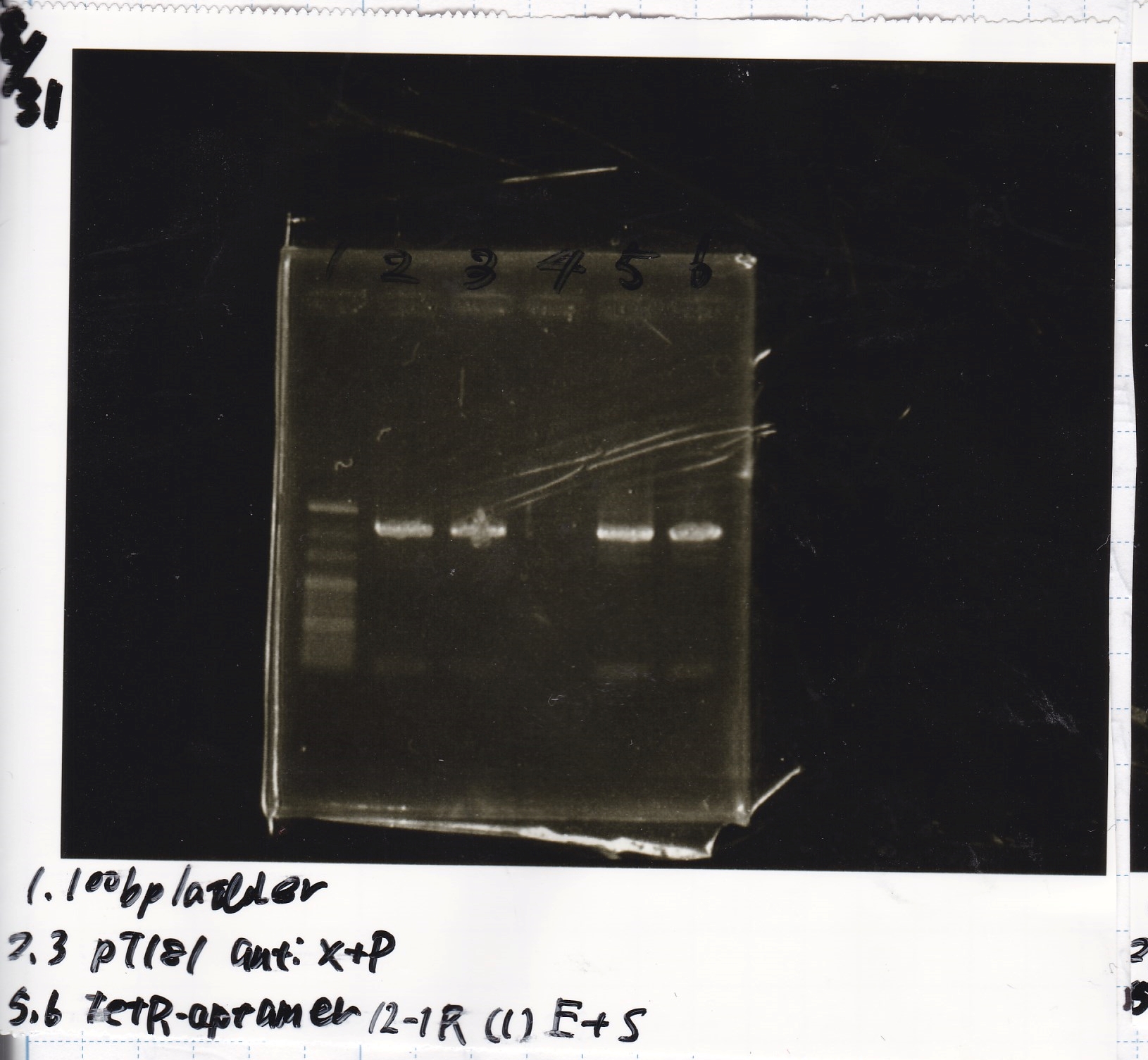
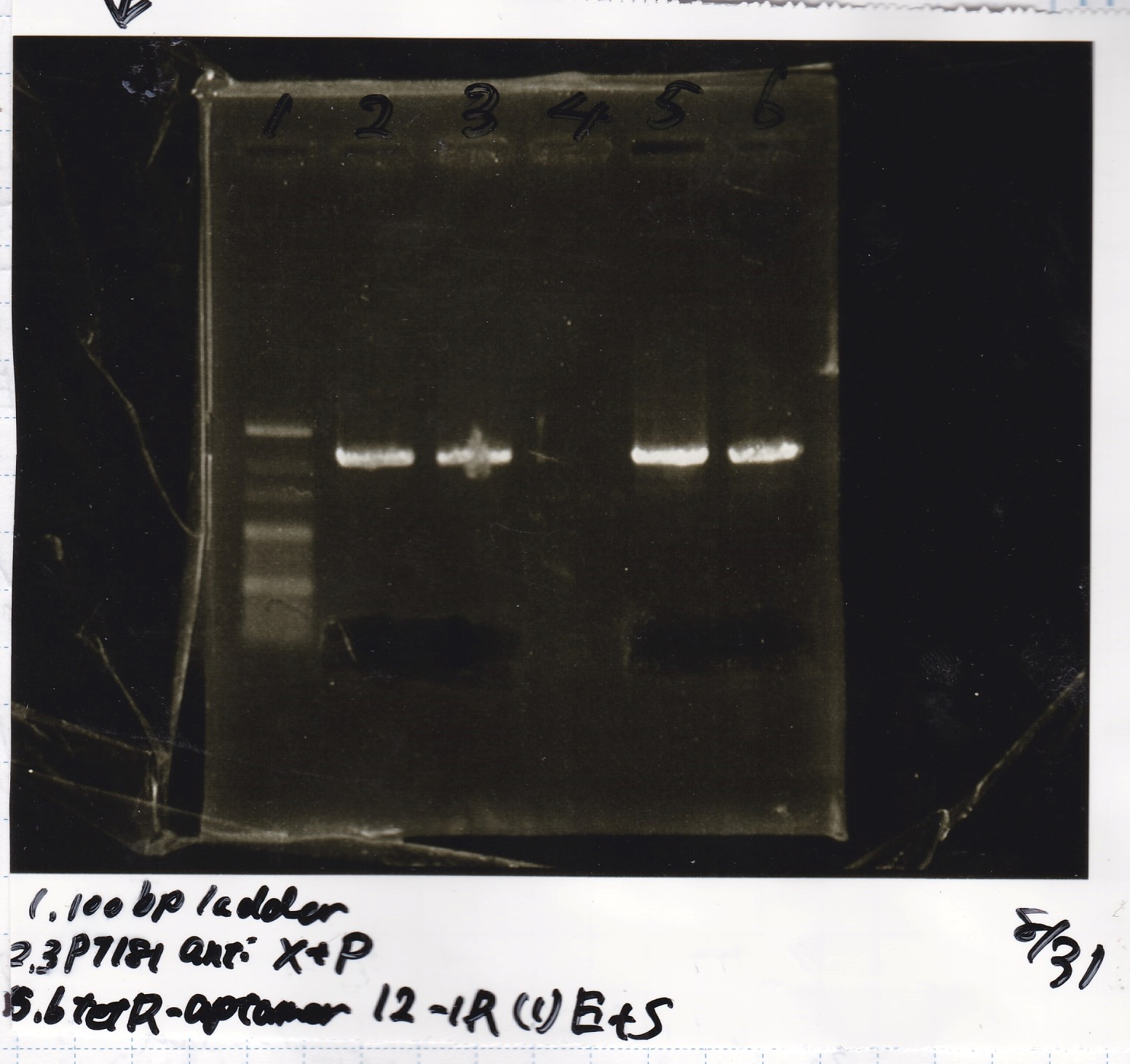
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-lysis2(EcoRI&SpeI) | 4.0 | 2.69 | 0.24
|
| pSB1C3(EcoRI&SpeI) | 6.0 | 2.48 | 0.05
|
| pSB1C3(XbaI&PstI) | -- | -- | --
|
| tRNA-Spinach | -- | -- | --
|
| pT181 attenuator-(2)(EcoRI&SpeI) | -- | -- | --
|
| pT181 anttenuator-(2)(SbaI&PstI) | -- | -- | --
|
| pT181 antisense-(2)(EcoRI&SpeI) | -- | -- | --
|
| pT181 antisense-(2)(XbaI&PstI) | -- | -- | --
|
| tetR aptamer 12_1R(EcoRI&SpeI) | -- | -- | --
|
Ligation
Nakamoto and Hirano
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 8/30 pSB1C3 (EcoRI & SpeI) | 6.6 | 8/30 tRNA-Spinach (EcoRI & SpeI) | 12.5 | 3.5
|
| experiment | 8/30 pSB1C3 (EcoRI & SpeI) | 6.6 | 8/30 pT181 antisense (EcoRI & SpeI) | 12.5 | 3.5
|
| experiment | 8/30 pSB1C3 (XbaI & Pst) | 12.5 | 8/30 pT181 attenuator (XbaI & PstI) | 25 | 3.5
|
| experiment | 8/30 pSB1C3 (XbaI & Pst) | 12.5 | 8/30 pT181 antisense (XbaI & PstI) | 25 | 3.5
|
| experiment | 8/20 DT (EcoRI & XbaI) | 6.6 | 8/30 pT181 attenuator(2) (EcoRI & SpeI) | 12.5 | 3.5
|
| experiment | 8/20 DT (EcoRI & XbaI) | 6.6 | 8/30 pT181 antisense (EcoRI & SpeI) | 12.5 | 3.5
|
| experiment | 8/27 DT (EcoRI & XbaI) | 3.0 | 8/30 tRNA-Spinach (EcoRI & SpeI) | 12.5 | 3.5
|
| experiment | 8/20 DT (EcoRI & XbaI) | 6.6 | 8/30 RBS-lysis2 (EcoRI & SpeI) | 12.5 | 3.5
|
| experiment | 8/22 J23100 (SpeI & PstI) | -- | RBS-tetR-DT (XbaI & PstI) | -- | 3.5
|
| experiment | 8/22 Plac (SpeI & PstI) | 8.0 | 8/18 RBS-lacZα-DT (XbaI & PstI) | 10 | 3.5
|
Samples were evaporeted used evaporator into about 7 µL
Miniprep
Kojima and Nakamoto
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 8/30 Plux-RBS-GFP-DT-(1) | 124 | 1.77 | 1.87
|
| 8/30 Plux-RBS-GFP-DT-(2) | 213 | 1.75 | 1.52
|
| 8/30 Pbad/araC-RBS-RFP | 427 | 1.70 | 1.56
|
| 8/30 Pbad/araC-RBS-RFP | 670 | 1.30 | 0.83
|
| 8/30 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(1) | 332 | 1.73 | 2.04
|
| 8/30 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(2) | 478 | 1.74 | 2.09
|
| 8/30 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(3) | 146 | 1.77 | 2.11
|
| 8/30 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT-(4) | 454 | 1.75 | 2.19
|
Sep 1
Transformation
No name
| Name | Sample | Competent Cells | Total | Plate
|
| 8/31 tRNA-Spinach(pSB1C3) | 2µL | 20µL | 22µL | CP
|
| 8/31 pT181 antisense(pSB1C3) | 2µL | 20µL | 22µL | CP
|
| 8/31 pT181 attenuator(pSB1C3) | 2µL | 20µL | 22µL | CP
|
| 8/31 pT181 antisense(pSB1C3) | 2µL | 20µL | 22µL | CP
|
| 8/31 pT181 attenuator(2)-DT | 2µL | 20µL | 22µL | CP
|
| 8/31 pT181 antisense-DT | 2µL | 20µL | 22µL | CP
|
| 8/31 tRNA-Spniach-DT | 2µL | 20µL | 22µL | CP
|
| 8/31 RBS-lysis2-DT | 2µL | 20µL | 22µL | CP
|
| 8/31 Pconst-RBS-tetR-DT | 2µL | 20µL | 22µL | Amp
|
| 8/31 Plac-RBS-lacZα-DT | 2µL | 20µL | 22µL | CP
|
| 8/31 Ptet-RBS-lacZα-DT | 2µL | 20µL | 22µL | CP
|
| 8/31 | 2µL | 20µL | 22µL | CP
|
ligation production PCR
No name
| Sample | base pair
|
| 8/31 tRNA Spinach(pSB1C3) | 459
|
| 8/31 pT181 antisense(2)(pSB1C3)(EcoRI+SpeI) | 415
|
| 8/31 pT181 attenuator(2)(pSB1C3)(XbaI+PstI) | 601
|
| 8/31 pT181 antisense(2)(pSB1C3)(XbaI+PstI) | 415
|
| 8/31 pT181 attenuator(2)-DT | 754
|
| 8/31 pT181 antisense(2)-DT | 577
|
| 8/31 tRNA Spinach-DT | 596
|
| 8/22 pSB1C3(2)(controll) | 1383
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30min | 30cycle
|

Ligation
Nakamoto
| state | Vector | Inserter
|
| experiment | DT(EcoRI+SpeI) | 5.5 | 8/28 RBS-lysis1(EcoRI+SpeI)2.4ng/µL | 2.4
|
1. Samples were evaporeted used evaporator into about 7 µL.
2. Add Ligation High 3.5µ:L and incubate 16°C 1hour.
Liquid Culture
Tatsui,Stephane,Nakamoto
| Sample | medium
|
| 8/21 pSB1C3(BBa_J04450)-(1) | Plusgrow medium(+CP)
|
| 8/21 pSB1C3(BBa_J04450)-(2) | Plusgrow medium(+CP)
|
| RBS-lysis3-DT | Plusgrow medium(+CP)
|
M9 Medium
No name
5xM9 liquid medium
| volume | 100mL
|
| NaHPO4 | 30g
|
| KH2PO | 3g
|
| Nacl | 0.25g
|
| NH4Cl | 0.5g
|
| 3.75% agar solution | 400mL
|
| 1M MgSO4 | 0.5mL
|
| 2M Glucose | 2.8mL
|
| 1% Vitamin B1 | 0.5mL
|
| 1M CaCl2 | 0.05mL
|
1. mix 5xM9 medium and 3.75%Agar solution 400mL
2. Add 1M MgSO4, 2M Glucose and 1 VitaminB1 0.5mL
3. Add CaCl2 while shaking Erlenmeyer flask.
Miniprep
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 8/21 pSB1C3(1) | 309.2 | 1.69 | 1.72
|
| 8/21 pSB1C3(2) | 201.6 | 1.65 | 1.76
|
Kanamycin applicaton
Hirano
Add 20µL Kanamycin into 80µL MilliQ and apply it on a 9/1 M9 plate
Plating
Hirano
Apply entA-colony liquid culture (100µL) on M9(Km)plate and incubate 37°C
2 fold diluted OD 600 2.107
Liquid Culture
Hirano
| Sample | medium
|
| 9/1 entA- Master Plate-(4) | SOB Medium(+Km)
|
Sep 2
Colony PCR
Tatsui
| Sample | base pair
|
| 9/1 RBS-lysis2-DT-(1) | 985
|
| Ptet-RBS-lacZα-DT-(1) | 765
|
| Plac-RBS-lacZα-DT-(1) | 765
|
| Plac-RBS-lacZα-DT-(2) | 765
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min | 30cycles
|
Electrophoresis
No name
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | 9/1 RBS-lysis2+DT -(1)
|
| 3 | 9/1 Ptet+RBS-lacZα-DT -(1)
|
| 4 | 9/1 Plac+RBS-lacZα-DT -(1)
|
| 5 | 9/1 Plac+RBS-lacZα-DT -(2)
|
| 6 | 100bp ladder
|
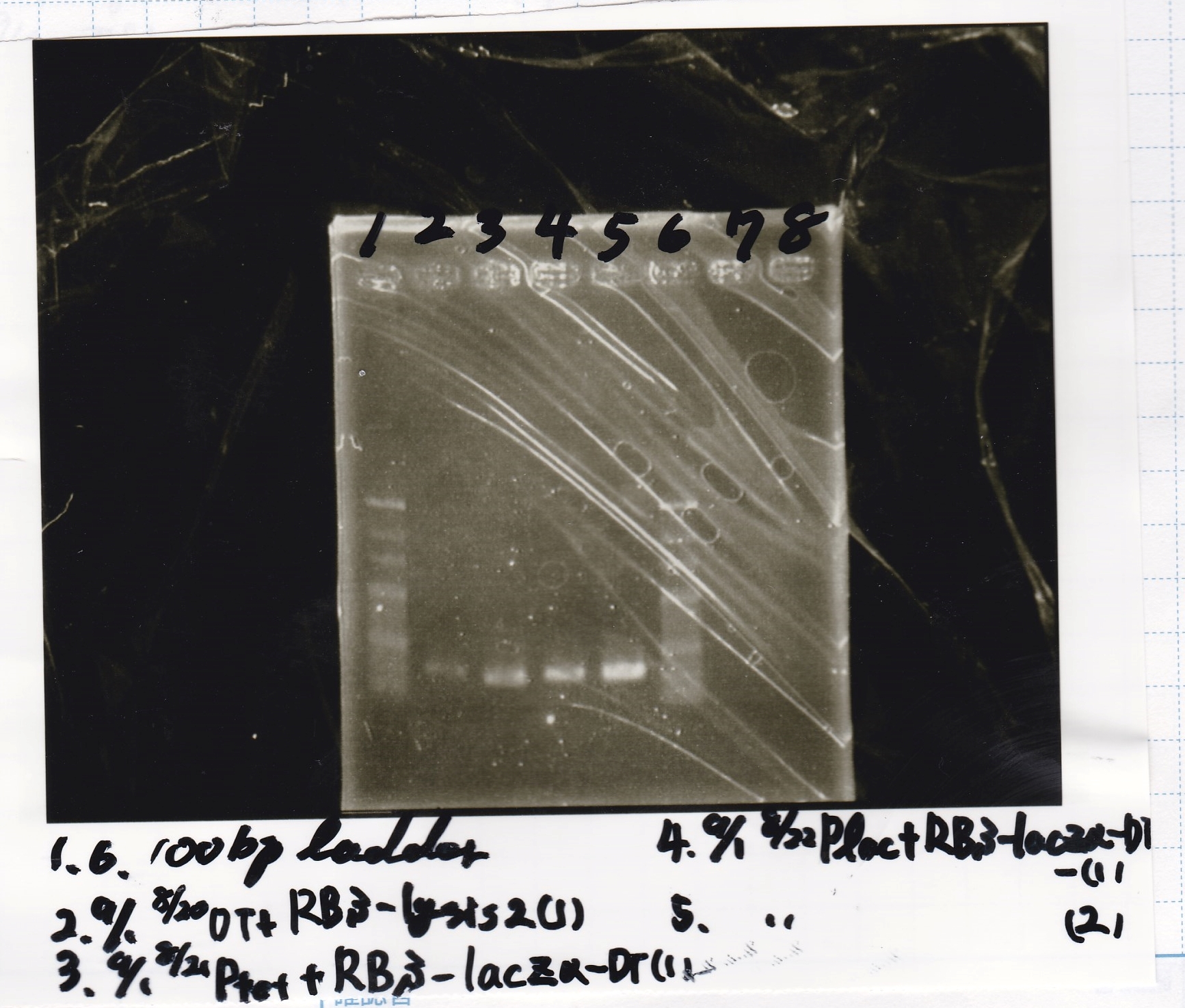
Restriction Enzyme Digestion
Nakamoto
| | pSB1C3-(1) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2 cuts(EcoRI+SpeI) | 7µL | 1µL | 1µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 17.7µL | 30µL
|
| NC(EcoRI+SpeI) | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.6µL | 10µL
|
| 2 cuts(XbaI+PstI) | 7µL | 0µL | 0µL | 1µL | 1µL | 0µL | 3µL | 0.3µL | 17.7µL | 30µL
|
| NC(XbaI+PstI) | 0.3µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.6µL | 10µL
|
| | 8/21 tRMA-spinach(1) | EcoRI | SpeI | BufferB | BSA | MilliQ | total
|
| 2 cuts | 8µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 17.6µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
| | 8/21 tetR aptamer12_1R-(1) | EcoRI | SpeI | BufferB | BSA | MilliQ | total
|
| 2 cuts | 6µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 18.7µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 0.1µL | 8.5µL | 10µL
|
| | 8/21 pT181 attenuator-(2) | EcoRI | SpeI | XbaI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2 cuts(EcoRI+SpeI) | 8µL | 1µL | 1µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 16.3µL | 30µL
|
| NC(EcoRI+SpeI) | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.4µL | 10µL
|
| 2 cuts(XbaI+PstI) | 8µL | 0µL | 0µL | 1µL | 1µL | 0µL | 3µL | 0.3µL | 16.3µL | 30µL
|
| NC(XbaI+PstI) | 0.5µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
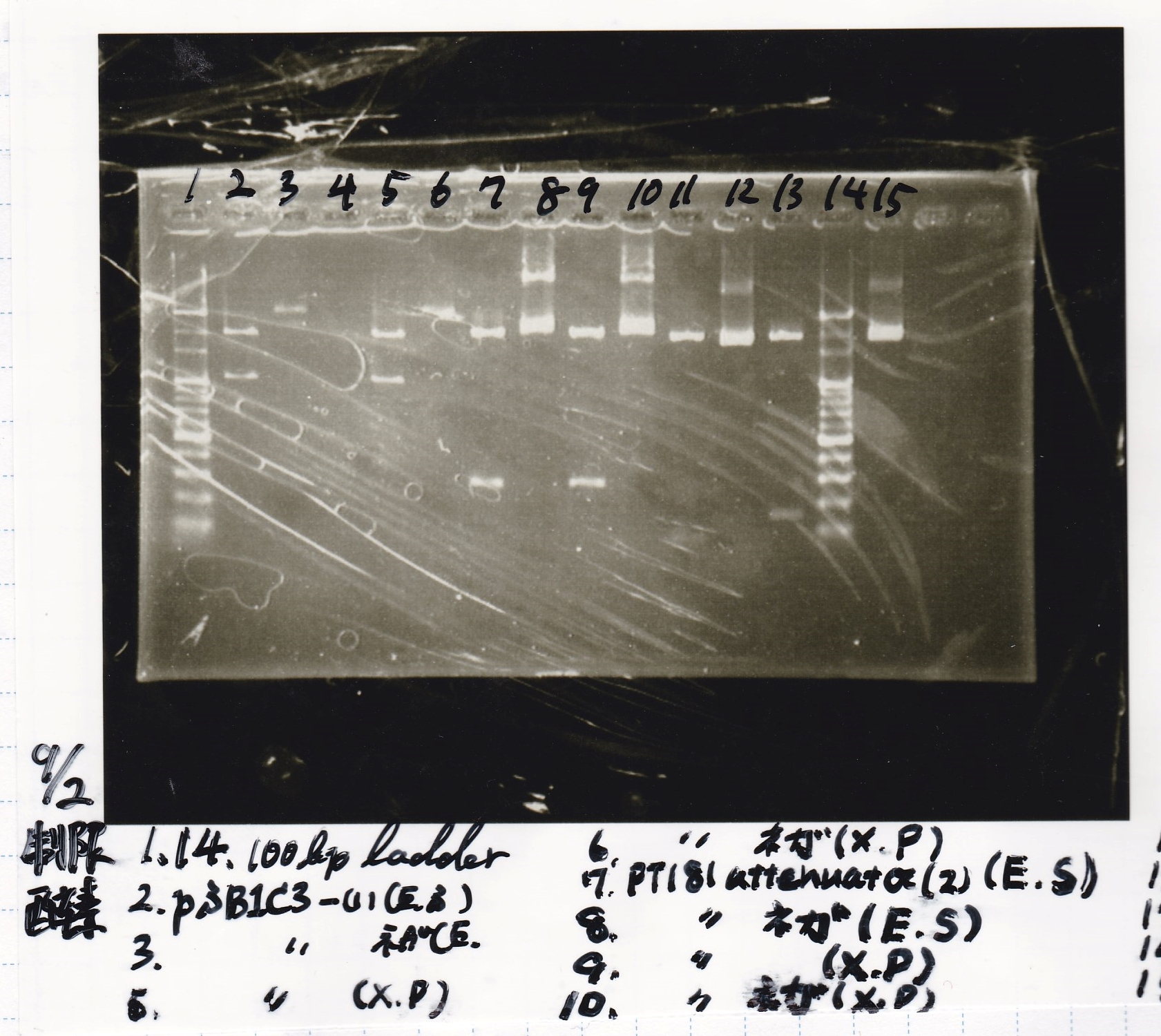
Liquid Culture
Kojima
| Sample | medium
|
| tRNA-Spinach-1 | Plusgrow medium(+CP)
|
| tetR aptamaer12_1R-1 | Plusgrow medium(+CP)
|
| pT181 attenuator-1 | Plusgrow medium(+CP)
|
| pT181 antisense-1 | Plusgrow medium(+CP)
|
Master Plate
Kojima
| Number | Use LB plate(+CP)
|
| 1 | tRNA Spinach-1
|
| 2 | tetR aptamaer12_1R
|
| 3 | tetR aptamaer12_P
|
| 4 | tetR aptamaer12_1M
|
| 5 | pT181 attenuator-2
|
| 6 | Fusion1 attenuator-1
|
| 7 | Fusion3m2 attenuator-1
|
| 8 | pT181 antisense-1
|
| 9 | Fusion1 antisense-1
|
| 10 | Fuaion6 antisense-1
|
Electrophoresis
Nakamoto & Tatsui
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | pSB1C3 -(1) | EcoRI | SpeI
|
| 3 | pSB1C3 -(1) | -- | --
|
| 5 | pSB1C3 -(1) | XbaI | PstI
|
| 6 | pSB1C3 -(1) | -- | --
|
| 7 | pT181 attenuator (2) | EcoRI | SpeI
|
| 8 | pT181 attenuator (2) | -- | --
|
| 9 | pT181 attenuator (2) | XbaI | SpeI
|
| 10 | pT181 attenuator (2) | -- | --
|
| 11 | tetR-aptamer 12_1R | EcoRI | SpeI
|
| 12 | tetR-aptamer 12_1R | -- | --
|
| 13 | tRNA Spinach (1) | EcoRI | SpeI
|
| 14 | 100bp ladder | -- | --
|
| 15 | tRNA Spinach (1) | -- | --
|

No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | -
|
| 2 | pSB1C3(EcoRI+SpeI) | EcoRI&SpeI
|
| 3
|
| 4 | -- | --
|
| 5 | 8/24 tetR aptamer 12_1R | EcoRI&SpeI
|
| 6
|


| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | -
|
| 2 | pT181 attenuator(2)(EcoRI+SpeI) | EcoRI&SpeI
|
| 3
|
| 4 | -- | --
|
| 5 | pT181 attenuator(2)(XbaI+PstI) | EcoRI&SpeI
|
| 6
|
| 7 | -- | --
|
| 8 | pT181 attenuator(2)(XbaI+PstI) | EcoRI&SpeI
|
| 9
|
| 10 | -- | --
|
| 11 | Spinach | EcoRI&SpaI
|
| 12
|


| Name | concentration[µg/mL] | 260/280 | 260/230
|
| pSB1C3 (EcoRI&SpeI) | 3.0 | 2.18 | 0.31
|
| pSB1C3 (XbaI&PstI) | 4.7 | 2.25 | 0.36
|
| pT181 attenuator-(2)(EcoR&SpeI) | 8.6 | 2.74 | 0.01
|
| pT181 attenuator-(2) (XbaI&PstI) | 16.5 | 2.46 | 0.03
|
| Spinach (EcoRI&SpeI) | 2.8 | 2.98 | 0.27
|
| tetR aptamer12_1R (EcoRI&SpeI) | 50.4 | 28.07 | 1.97
|
Colony PCR
No name
| Sample | base pair
|
| 9/1 Spinach(pSB1C3) | about 450
|
| 9/1 Spinach-DT | 596
|
| 9/1 RBS-lysis1-DT | 613
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 40s | 30cycles
|
| Sample | base pair
|
| 9/1 Ptet-RBS-lacZα-DT-2 | 738
|
| 9/1 Ptet-RBS-lacZα-DT-3 | 738
|
| 9/1 Ptet-RBS-lacZα-DT-4 | 738
|
| 9/1 Ptet-RBS-lacZα-DT-5 | 738
|
| 9/1 Ptet-RBS-lacZα-DT-6 | 738
|
| 9/1 RBS-lysis2-DT-2 | 985
|
| 9/1 RBS-lysis2-DT-3 | 985
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min | 30cycles
|

Electrophoresis
No name
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Spinach(pSB1C3)
|
| 3 | Spinach-DT
|
| 4 | RBS-lysis1-DT
|
| 5 | 100bp ladder
|

Master Plate
No name
| Number | Use LB plate(+CP)
|
| 1 | 9/1 Spinach(pSB1C3)
|
| 2 | 9/1 Spinach-DT-(1)
|
Liquid Culture
No name
| Sample | medium
|
| pT181 attenuator | Plusgrow medium (+CP)
|
| pT181 antisense | Plusgrow medium (+CP)
|
| Spinach | Plusgrow medium (+CP)
|
- incubated at 37 °C for 1 hour
Transformation
No name
| Name | Sample | Competent Cells | Total | Plate
|
| 8/28 RBS-lysis3-DT | 3µL | 30µL | 33µL | CP
|
Sep 3
Electrophoresis
Kojima
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | pT181 attenuator(2)-DT2(738)
|
| 3 | Ptet-RBS-lacZα-DT-2(756)
|
| 4 | Ptet-RBS-lacZα-DT-3
|
| 5 | Ptet-RBS-lacZα-DT-4
|
| 6 | Ptet-RBS-lacZα-DT-5
|
| 7 | Ptet-RBS-lacZα-DT-6
|
| 8 | RBS-lysis2-DT-2(985)
|
| 9 | RBS-lysis2-DT-3
|
| 10 | 100bp ladder
|
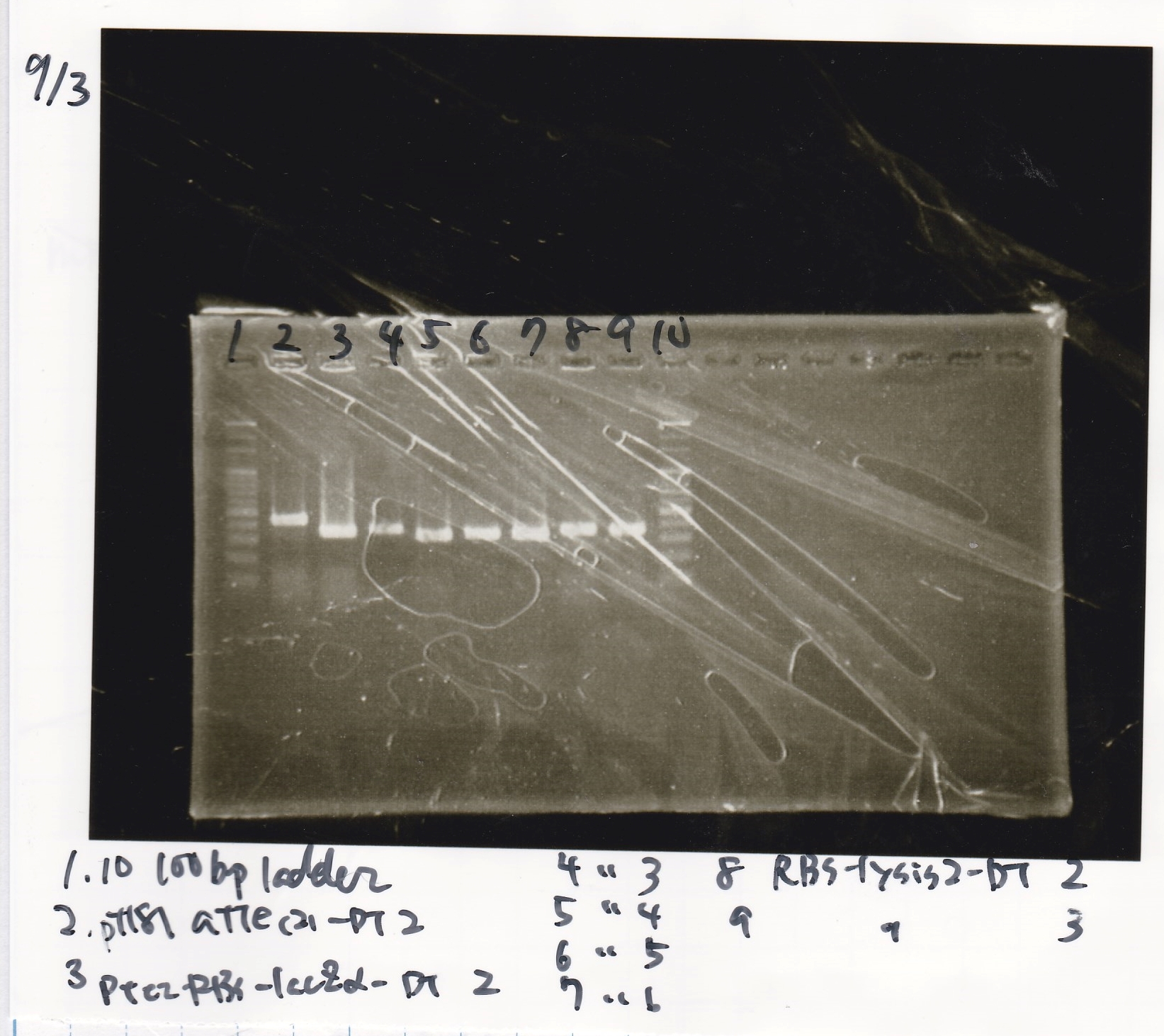
Miniprep
Tatsui
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/2 14:00 pT181 attenuator | 418.4 | 1.82 | 1.80
|
| 9/2 14:00 pT181 antisense | 362.2 | 1.92 | 1.96
|
| 9/2 14:00 Spinach | 438.3 | 1.86 | 1.99
|
| 9/2 14:00 tetR-apt 12_1R | 567.5 | 1.90 | 1.99
|
| 9/2 21:00 pT181 attenuator | 320.9 | 1.90 | 2.15
|
| 9/2 21:00 Spinach | 457.0 | 1.88 | 2.24
|
Liquid Culture
Kojima
| Sample | medium
|
| Spinach-DT(9/2 master plate) | plusgrow(CP)4ml
|
Restriction Enzyme Digestion
Kojima
| | DNA | EcoRI | SpeI | XbaI | PstI | 10xBuffer B(EcoRI&SpeI) | 10xBuffer D(XbaI&PstI) | 100xBSA | MilliQ | total
|
| 9/3 pT181attenuator 14:00 418.4ng/µL | 4.8µL | 1.0µL | 1.0µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 19.9µL | 30µL
|
| 9/3 pT181attenuator 14:00 418.4ng/µL | 0.2µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.7µL | 10µL
|
| 9/3 pT181attenuator 14:00 418.4ng/µL | 4.8µL | 0µL | 0µL | 1.0µL | 1.0µL | 0µL | 3µL | 0.3µL | 19.9µL | 30µL
|
| 9/3 pT181attenuator 14:00 418.4ng/µL | 0.2µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.7µL | 10µL
|
| | DNA | EcoRI | SpeI | XbaI | PstI | 10xBuffer B(EcoRI&SpeI) | 10xBuffer D(XbaI&PstI) | 100xBSA | MilliQ | total
|
| 9/3 pT181antisense 14:00 362.2ng/µL | 5.5µL | 1.0µL | 1.0µL | 0µL | 0µL | 3µL | 0µL | 0.3µL | 19.2µL | 30µL
|
| 9/3 pT181attenuator 14:00 418.4ng/µL | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 0.1µL | 8.6µL | 10µL
|
| 9/3 pT181attenuator 14:00 418.4ng/µL | 5.5µL | 0µL | 0µL | 1.0µL | 1.0µL | 0µL | 3µL | 0.3µL | 19.2µL | 30µL
|
| 9/3 pT181attenuator 14:00 418.4ng/µL | 0.3µL | 0µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0.1µL | 8.6µL | 10µL
|
| | DNA | EcoRI | SpeI | 10xBuffer B | 100xBSA | MilliQ | total
|
| 9/3 apt 12_1R 14:00 507.5ng/µL | 3.9µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 20.8µL | 30µL
|
| 9/3 apt 12_1R 14:00 507.5ng/µL | 0.2µL | 0µL | 0µL | 1µL | 0.1µL | 8.7µL | 10µL
|
| | DNA | EcoRI | XbaI | 10xBuffer D | 100xBSA | MilliQ | total
|
| 8/29 DT 166.2ng/µL | 12µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 12.7µL | 30µL
|
| 8/29 DT 166.2ng/µL | 0.6µL | 0µL | 0µL | 1µL | 0.1µL | 8.3µL | 10µL
|
| | DNA | EcoRI | SpeI | 10xBuffer B | 100xBSA | MilliQ | total
|
| 8/22 RBS-lysis1(2) 96ng/µL | 21µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 3.7µL | 30µL
|
| 8/22 RBS-lysis1(2) 96ng/µL | 1.0µL | 0µL | 0µL | 1µL | 0.1µL | 7.9µL | 10µL
|
| | DNA | EcoRI | SpeI | 10xBuffer B | 100xBSA | MilliQ | total
|
| 8/30 RBS-lysis2 220ng/µL | 9.1µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 15.6µL | 30µL
|
| 8/30 RBS-lysis2 220ng/µL | 0.5µL | 0µL | 0µL | 1µL | 0.1µL | 8.4µL | 10µL
|
| | DNA | EcoRI | SpeI | 10xBuffer B | 100xBSA | MilliQ | total
|
| 8/20 RBS-lysis3(1) 282ng/µL | 7.1µL | 1.0µL | 1.0µL | 3µL | 0.3µL | 17.6µL | 30µL
|
| 8/20 RBS-lysis3(1) 282ng/µL | 0.4µL | 0µL | 0µL | 1µL | 0.1µL | 8.5µL | 10µL
|
PCR
Hirano
| Plac(1) plasmid 153.7ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer (KaiA_BsaI_Mut_fwd) | primer (KaiA_BsaI_Mut_rer) | KOD plus-ver.2 | MilliQ | total
|
| 0.13 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16.37 | 25
|
| KaiB (plasmid) 20.2ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer (GG2_RBS_KaiB_fwd) | primer (KaiB_GG3_rer) | KOD plus-ver.2 | MilliQ | total
|
| 1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 15.5 | 25
|
| KaiC (plasmid) 4.4ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer | primer | KOD plus-ver.2 | MilliQ | total
|
| 1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 15.5 | 25
|
| DT-1 (plasmid) 153.7ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer | primer | KOD plus-ver.2 | MilliQ | total
|
| 0.1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16.4 | 25
|
| PKaiBC(PCR product) 16.3ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer | primer | KOD plus-ver.2 | MilliQ | total
|
| 0.7 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 15.8 | 25
|
| SasA(PCR product) 21.6ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer | primer | KOD plus-ver.2 | MilliQ | total
|
| 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16 | 25
|
| RpaA(PCR product) 15.4ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer | primer | KOD plus-ver.2 | MilliQ | total
|
| 0.6 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 15.9 | 25
|
| RpaB(PCR product) 26.7ng/µL | 10x buffer KOD plus-ver.2 | 2mM dNTPs | 25mM MgSO4 | primer | primer | KOD plus-ver.2 | MilliQ | total
|
| 0.4 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 16.1 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | | 68°C | --
|
| 5min | 10sec | 30sec | | 30
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | | 68°C | --
|
| 5min | 10sec | 30sec | | 30
|
Mutatation PCR
No name
| KaiABC(12.6pg/µL) | Primer(Kai_BsaIMut_fwd) | Primer(Kai_BsaIMut_rer) | Prime STAR Max Premix | MilliQ | total
|
| 1 | 1 | 1 | 25 | 22 | 50
|
| Primer(Kai_BsaIMut_fwd) | Primer(Kai_BsaIMut_rer) | Prime STAR Max Premix | MilliQ | total
|
| | 1 | 1 | 25 | 23 | 50
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 65°C | 72°C | --
|
| 5min | 10sec | 15sec | 40sec | 30
|
Colony PCR
| Sample | base pair
|
| 9/1 pT181antisense(pSB1C3) | 400bp
|
| 9/1 pT181antisense-DT | 547bp
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5m | 30s | 30s | 30s | 30
|
Electrophoresis
Kojima
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | 9/1 pT181antisense(pSB1C3)
|
| 3 | 9/1 pT181antisense
|
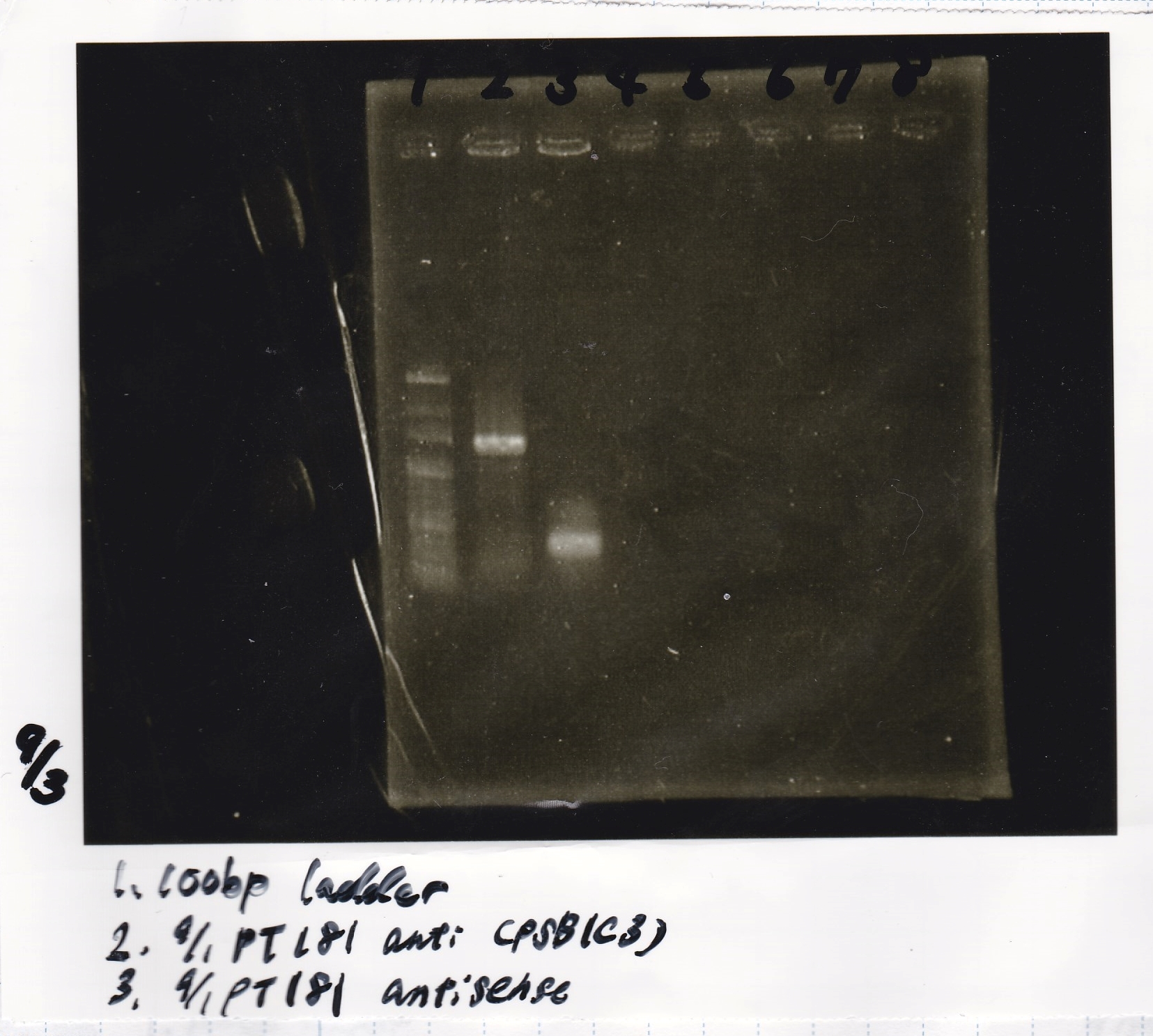
Electrophoresis
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | attenuator EcoRI&SpeI
|
| 3 | attenuator negative EcoRI&SpeI
|
| 4 | attenuator XbaI&PstI
|
| 5 | attenuator negative XbaI&PstI
|
| 6 | antisense EcoRI&SpeI
|
| 7 | antisense negative EcoRI&SpeI
|
| 8 | 100bp ladder
|
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | antisense XbaI&PstI
|
| 3 | antisense negative XbaI&PstI
|
| 4 | apt EcoRI&SpeI
|
| 5 | apt negative EcoRI&SpeI
|
| 6 | DT EcoRI&XbaI
|
| 7 | DT negative EcoRI&XbaI
|
| 8 | 100bp ladder
|

| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | RBS-lysis1 EcoRI&SpeI
|
| 3 | RBS-lysis1 negative
|
| 4 | RBS-lysis2 EcoRI&SpeI
|
| 5 | RBS-lysis2 negative
|
| 6 | RBS-lysis3 EcoRI&SpeI
|
| 7 | RBS-lysis3 negative
|
| 8 | 100bp ladder
|

| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 3 | pT181 attenuator | EcoRI&SpeI
|
| 4 | pT181 attenuator | EcoRI&SpeI
|
| 6 | pT181 attenuator | XbaI&PstI
|
| 7 | pT181 attenuator | XbaI&PstI
|
| 9 | pT181 antisense | EcoRI&SpeI
|
| 10 | pT181 antisense | EcoRI&SpeI
|

| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 2 | pT181 antisense | XbaI&PstI
|
| 3 | pT181 antisense | XbaI&PstI
|
| 5 | apt12_1R | EcoRI&SpeI
|
| 6 | apt12_1R | EcoRI&SpeI
|
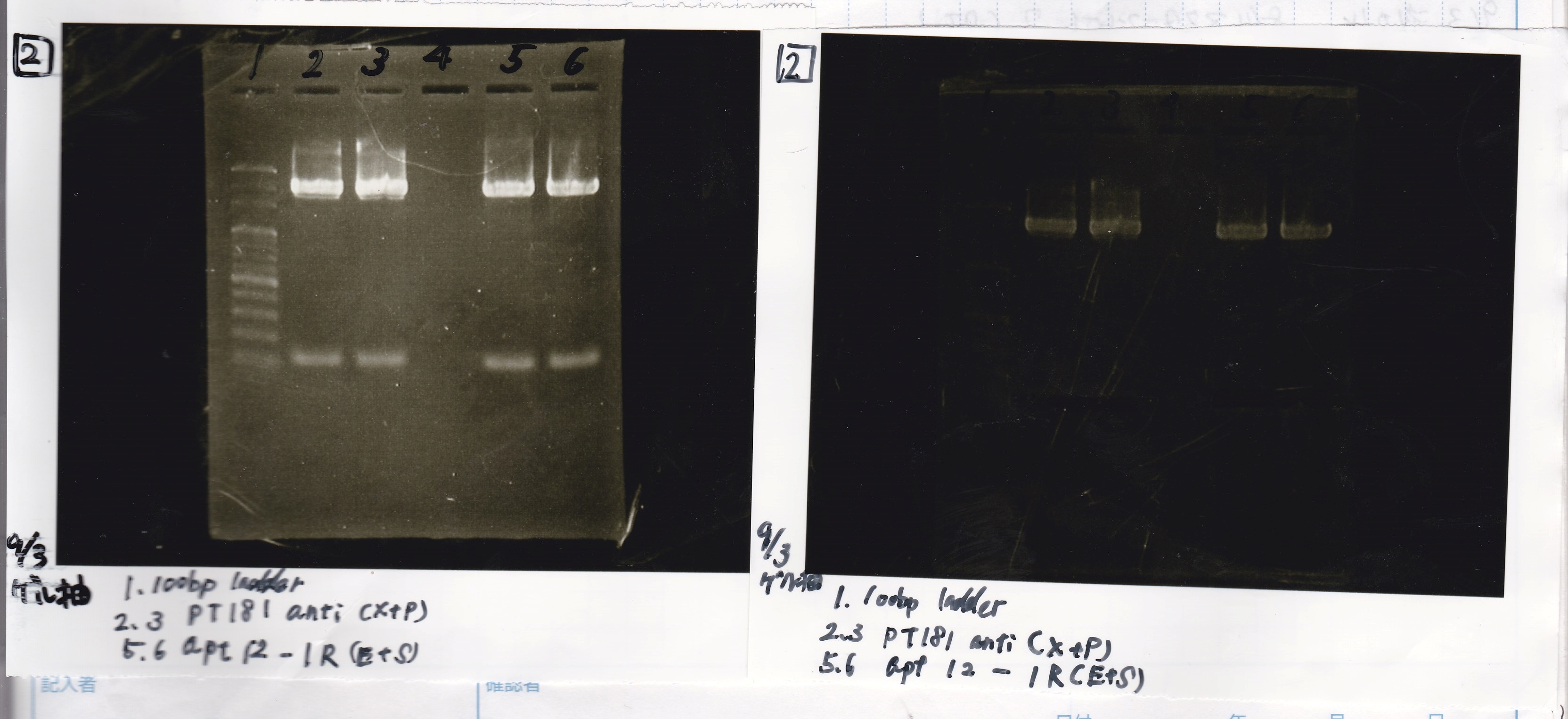
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 2 | DT | EcoRI&XbaI
|
| 3 | DT | EcoRI&XbaI
|
| 5 | RBS-lysis1 | EcoRI&SpeI
|
| 6 | RBS-lysis1 | EcoRI&SpeI
|
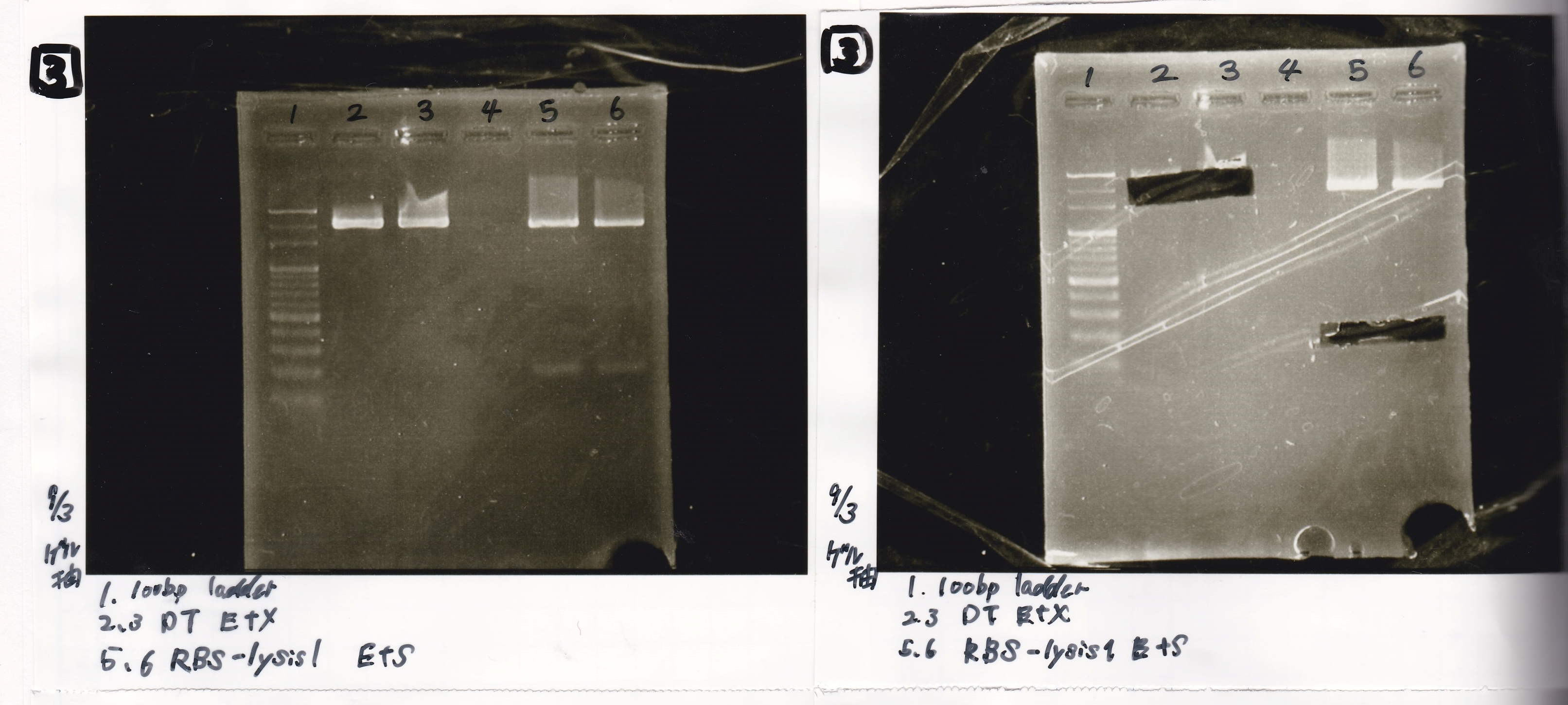
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 2 | RBS-lysis2 | EcoRI&SpeI
|
| 3 | RBS-lysis2 | EcoRI&SpeI
|
| 5 | RBS-lysis3 | EcoRI&SpeI
|
| 6 | RBS-lysis3 | EcoRI&SpeI
|

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| pT181 attenuator(EcoRI&SpeI) | 10.0 | 1.86 | 0.39
|
| pT181 attenuator(XbaI&PstI) | 11.7 | 1.65 | 0.36
|
| pT181 antisense(EcoRI&SpeI) | 4.2 | 1.95 | 0.25
|
| pT181 antisense(XbaI&PstI) | 6.6 | 1.78 | 0.33
|
| tRNA 12_1R apt(EcoRI&SpeI) | 5.7 | 1.73 | 0.29
|
| DT(EcoRI&XbaI) | 16.6 | 1.99 | 0.76
|
| RBS-lysis1(EcoRI&SpeI) | 8.8 | 1.65 | 0.32
|
| RBS-lysis2(EcoRI&SpeI) | 13.4 | 1.61 | 0.48
|
| RBS-lysis3(EcoRI&SpeI) | 10.2 | 2.19 | 0.51
|
Liquid Culture
No name
| Sample | medium
|
| 8/16master plate7(DT) |
|
| 8/21RBS-lysis1 |
|
Sep 4
Ligation
No name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/3 DT(EcoRI&XbaI) | 3.2µL | 9/3 RBS-lysis1 | 2.4µL | 2.7µL
|
| experiment | 9/3 DT(EcoRI&XbaI) | 3.0µL | 9/3 RBS-lysis2 | 4.7µL | 4.0µL
|
| experiment | 9/3 DT(EcoRI&XbaI) | 3.0µL | 9/3 RBS-lysis3 | 8.8µL | 4.0µL
|
| experiment | 9/3 DT(EcoRI&XbaI) | 3.0µL | 9/3 aptamer12_1R(EcoRI&SpeI) | 1.9µL | 2.45µL
|
| experiment | 9/2 pSB1C3(XbaI&PstI) | 10.6µL | 9/3 pT181attenuator (XbaI& PstI) | 3.2µL | 4.0µL
|
| experiment | 9/2 pSB1C3(XbaI&PstI) | 9.6µL | 9/3 pT181antisense (XbaI& PstI) | 2.1µL | 4.0µL
|
| experiment | 9/2 pSB1C3(EcoRI&SpeI) | 16.7µL | 9/3 aptamer12_1R(EcoRI&SpeI) | 2.0µL | 4.0µL
|
incubate 16 °C 1 hour
Transformation
Tatsui
| Name | Sample | Competent Cells | Plate
|
| 9/4 RBS-lysis1-DT | 2µL | 20µL | Amp
|
| 9/4 RBS-lysis2-DT | 2µL | 20µL | Amp
|
| 9/4 RBS-lysis3-DT | 2µL | 20µL | Amp
|
| 9/4 aptamer12_1R-DT | 2µL | 20µL | Amp
|
| 9/4 pT181attenuator –pSB1C3 | 2µL | 20µL | Amp
|
| 9/4 pT181antisense-pSB1C3 | 2µL | 20µL | Amp
|
| 9/4 aptamer12_1R –pSB1C3 | 2µL | 20µL | Amp
|
| 9/4 pSB4K5(2013plate5 56) | 2µL | 20µL | Amp
|
| 9/3 Mutagenesis product 1(KaiABC) | 2µL | 20µL | Amp
|
| 9/3 Mutagenesis product 2(NC) | 2µL | 20µL | Amp
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| DT | 193.1 | 1.93 | 1.81
|
| Spinach-DT | 373.3 | 1.92 | 1.69
|
| RBS-lysis1 | 332.0 | 1.95 | 1.62
|
Master Plate
Hirano
| Number | Use LB plate (+CP)
|
| 1 | 8/10 Plac(CP)-2
|
| 2 | 8/10 Plac(CP)-2
|
| 3 | 8/10 Plac(CP)-2
|
| Number | Use LB plate (+Amp)
|
| 1 | 8/9 J23100(Amp)-1
|
| 2 | 8/9 J23100(Amp)-1
|
| 3 | 8/9 J23100(Amp)-1
|
Liquid Culture
Hirano
| Sample | medium
|
| 8/10 Plac(CP)-2 | Plusgrow medium(+CP)
|
| 8/9 J23100(Amp)-1 | Plusgrow medium(+Amp)
|
PCR
Hirano
| 8/29 Pλ-RBS-luxI-DT | KOD plus | 10x buffer | dNTP | MgSO4 | F primer (RBS-luxI-DT-cloning-fwd) | R primer(RBS-luxI-DT-cloning-rev) | MilliQ | total
|
| 0.3µL | 0.5µL | 2.5µL | 2.5µL | 1.5µL | 0.75µL | 0.75µL | 16.2µL | 25µL
|
| 8/31 Pbad/araC-RBS-RFP(1) | KOD plus | 10x buffer | dNTP | MgSO4 | F primer (Pbad/araC-pSB1C3-cloning-fond) | R primer(Pbad/araC-cloning-rex) | MilliQ | total
|
| 0.3µL | 0.5µL | 2.5µL | 2.5µL | 1.5µL | 0.75µL | 0.75µL | 16.2µL | 25µL
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 65°C | 68°C | --
|
| 5min | 10s | 30s | 3min10s | 30cycles
|
Restriction Enzyme Digestion
No name
| | 8/21 Pconst | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 10.5µL | 1.0µL | 1.0µL | 3.0µL | 0.3µL | 14.2µL | 30 µL
|
| NC | 0.5µL | 0µL | 0µ | 1.0µL | 0.1µL | 8.4µL | 10 µL
|
| | 8/29 Plac | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 13.0µL | 1.0µL | 1.0µL | 3.0µL | 0.3µL | 11.7µL | 30 µL
|
| NC | 0.7µL | 0µL | 0µ | 1.0µL | 0.1µL | 8.2µL | 10 µL
|
| | 8/20 Ptet | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 9.0µL | 1.0µL | 1.0µL | 3.0µL | 0.3µL | 15.7µL | 30 µL
|
| NC | 0.5µL | 0µL | 0µ | 1.0µL | 0.1µL | 8.4µL | 10 µL
|
| | 8/20 RBS-tetR-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 8.5µL | 1.0µL | 1.0µL | 3.0µL | 0.3µL | 16.2µL | 30 µL
|
| NC | 0.5µL | 0µL | 0µ | 1.0µL | 0.1µL | 8.4µL | 10 µL
|
| | spinach -DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 5.4µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 16.6µL | 30 µL
|
| NC | 0.3µL | 0µL | 0µ | 1.0µL | 1.0µL | 7.7µL | 10 µL
|
| | 8/17 RBS-lacZα-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 11µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 11µL | 30 µL
|
| NC | 0.5µL | 0µL | 0µ | 1.0µL | 1.0µL | 7.5µL | 10 µL
|
Transformation
No name
| Name | Sample | Competent Cells | Total | Plate
|
| Ptrc KaiC | 1µL | 10µL | 11µL | Amp
|
| Ptrc KaiB | 1µL | 10µL | 11µL | Amp
|
| pSΩ | 1µL | 10µL | 11µL | Amp
|
Electrophoresis
No name
1
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Pconst | SpeI | PstI
|
| 3 | Pconst NC | -- | --
|
| 4 | Plac | SpeI | PstI
|
| 5 | Plac NC | -- | --
|
| 6 | 100bp ladder | -- | --
|
2
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Ptet | SpeI | PstI
|
| 3 | Ptet NC | -- | --
|
| 4 | RBS-tetR-DT | XbaI | PstI
|
| 5 | RBS-tetR-DT NC | -- | --
|
| 6 | 100bp ladder | -- | --
|
3
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Spinach-DT | XbaI | PstI
|
| 3 | Spinach-DT NC | -- | --
|
| 4 | RBS-lacZα-DT | XbaI | PstI
|
| 5 | RBS-lacα-DT NC | -- | --
|
| 6 | 100bp ladder | -- | --
|
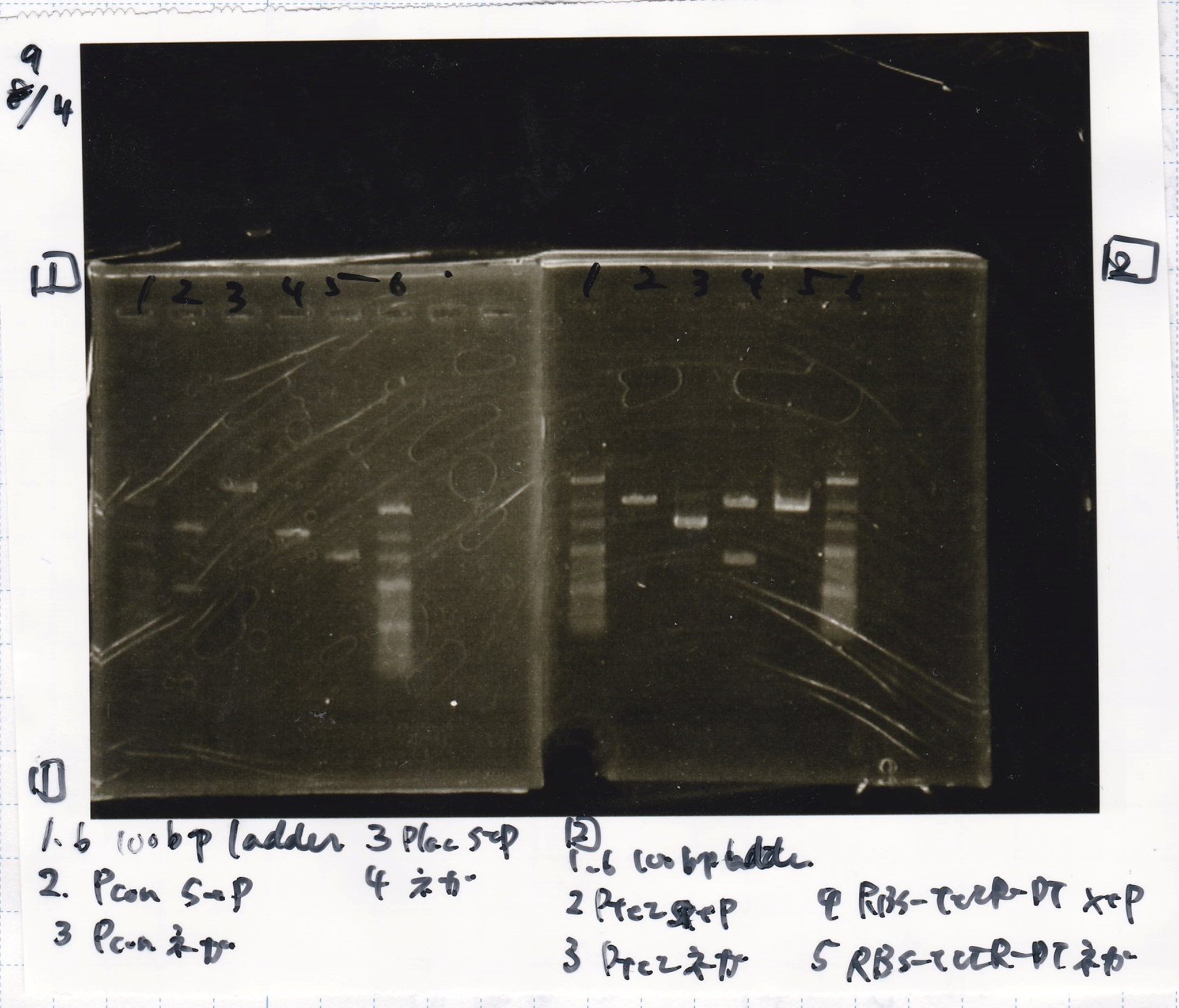
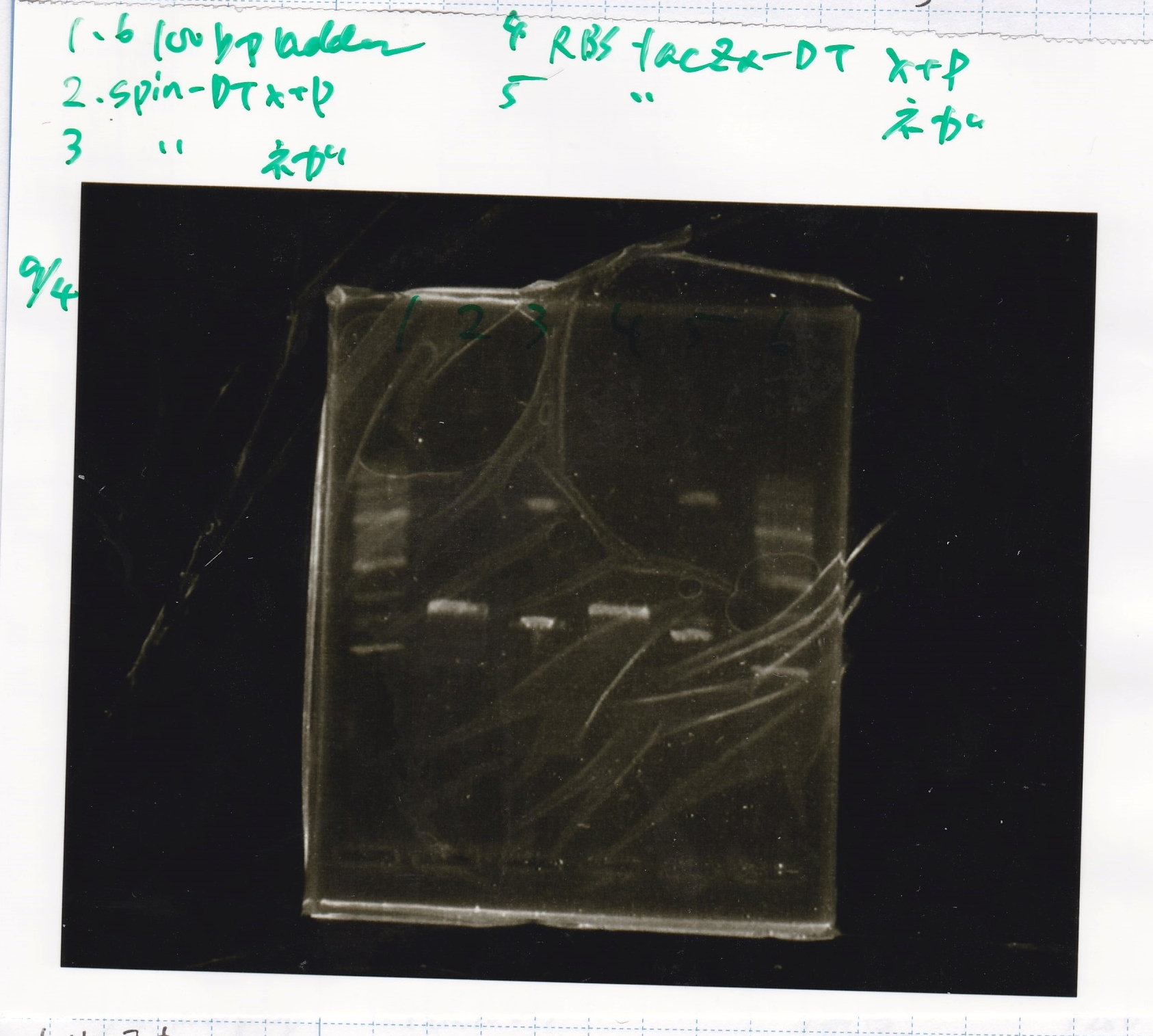
No name
1
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | Pcon | SpeI&PstI
|
| 3 | Pcon | SpeI&PstI
|
| 4 | -- | --
|
| 5 | Plac | SpeI&PstI
|
| 6 | Plac | SpeI&PstI
|
2
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | Ptet | SpeI&PstI
|
| 3 | Ptet | SpeI&PstI
|
| 4 | -- | --
|
| 5 | RBS-tetR-DT | XbaI&PstI
|
| 6 | RBS-tetR-DT | XbaI&PstI
|

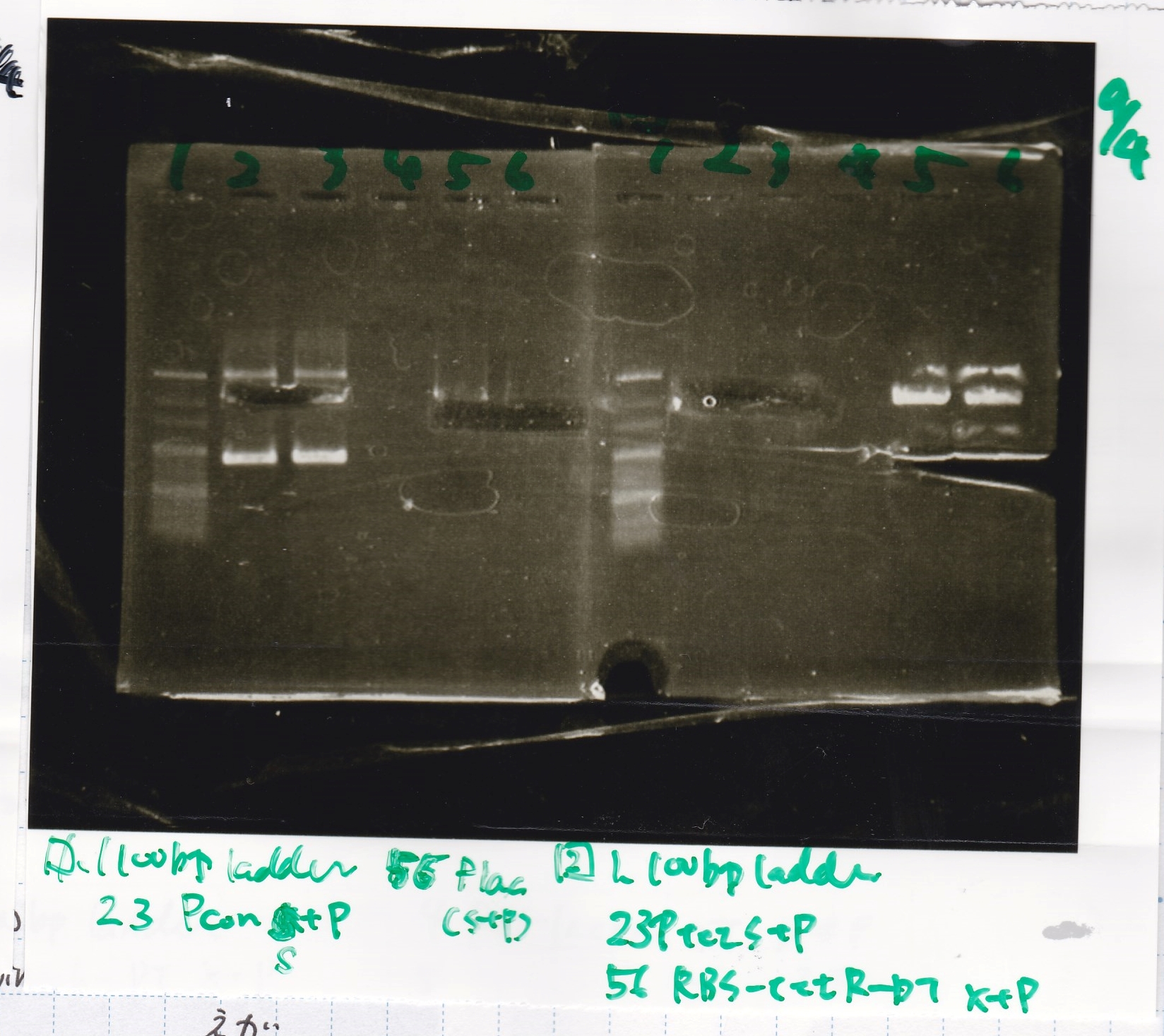
| Name | quantity | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon | -- | 13.2 | 1.99 | -1.64
|
| Plac | -- | 15.3 | 2.07 | -0.39
|
| Ptet | -- | 30.1 | 1.97 | -0.97
|
| RBS-tetR-DT | -- | 3.0 | 2.44 | 0.88
|
No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | spinach-DT | XbaI&PstI
|
| 3 | spinach-DT | XbaI&PstI
|
| 4 | -- | --
|
| 5 | RBS-lacZα-DT | XbaI&PstI
|
| 6 | RBS-lacZα-DT | XbaI&PstI
|
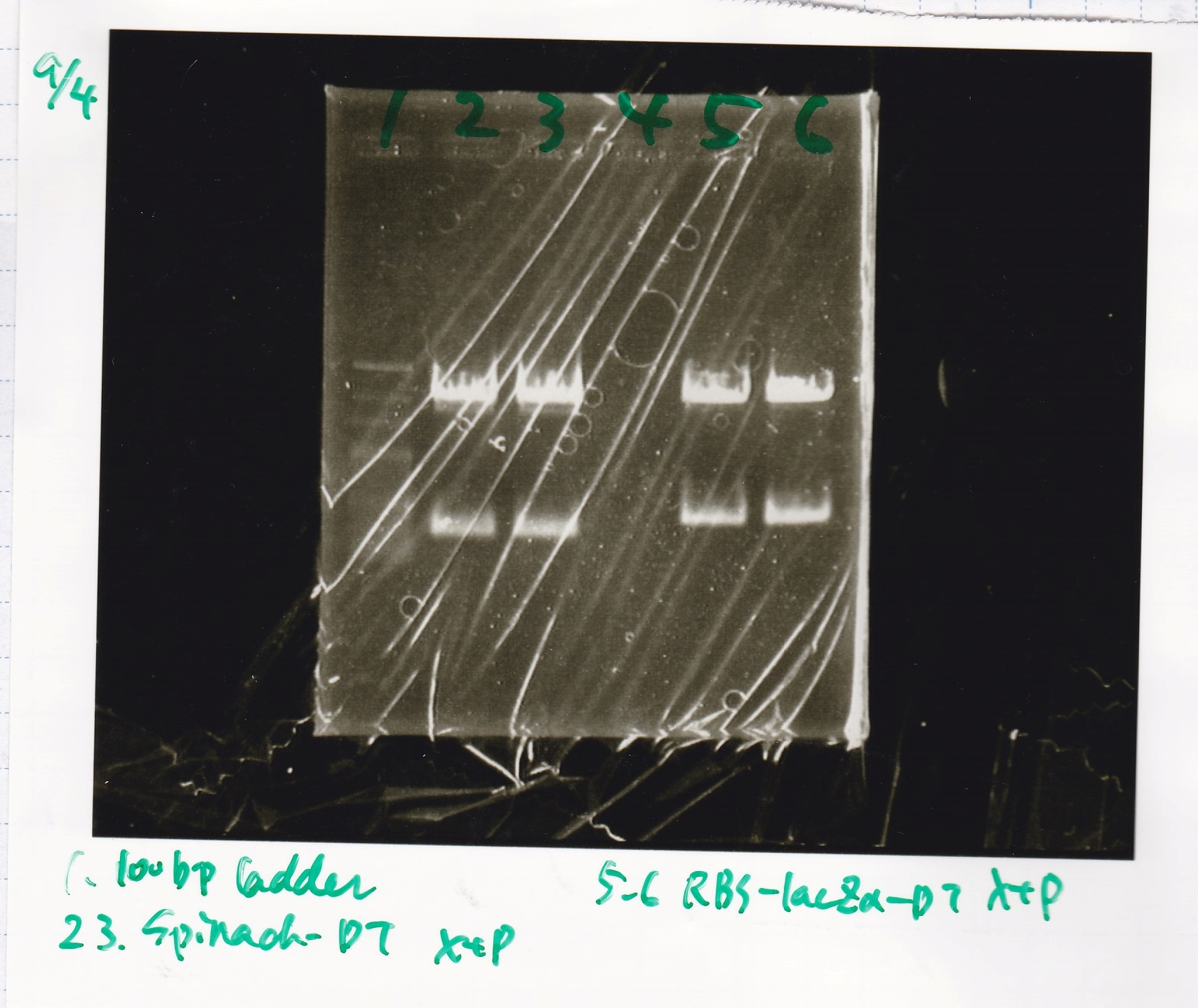
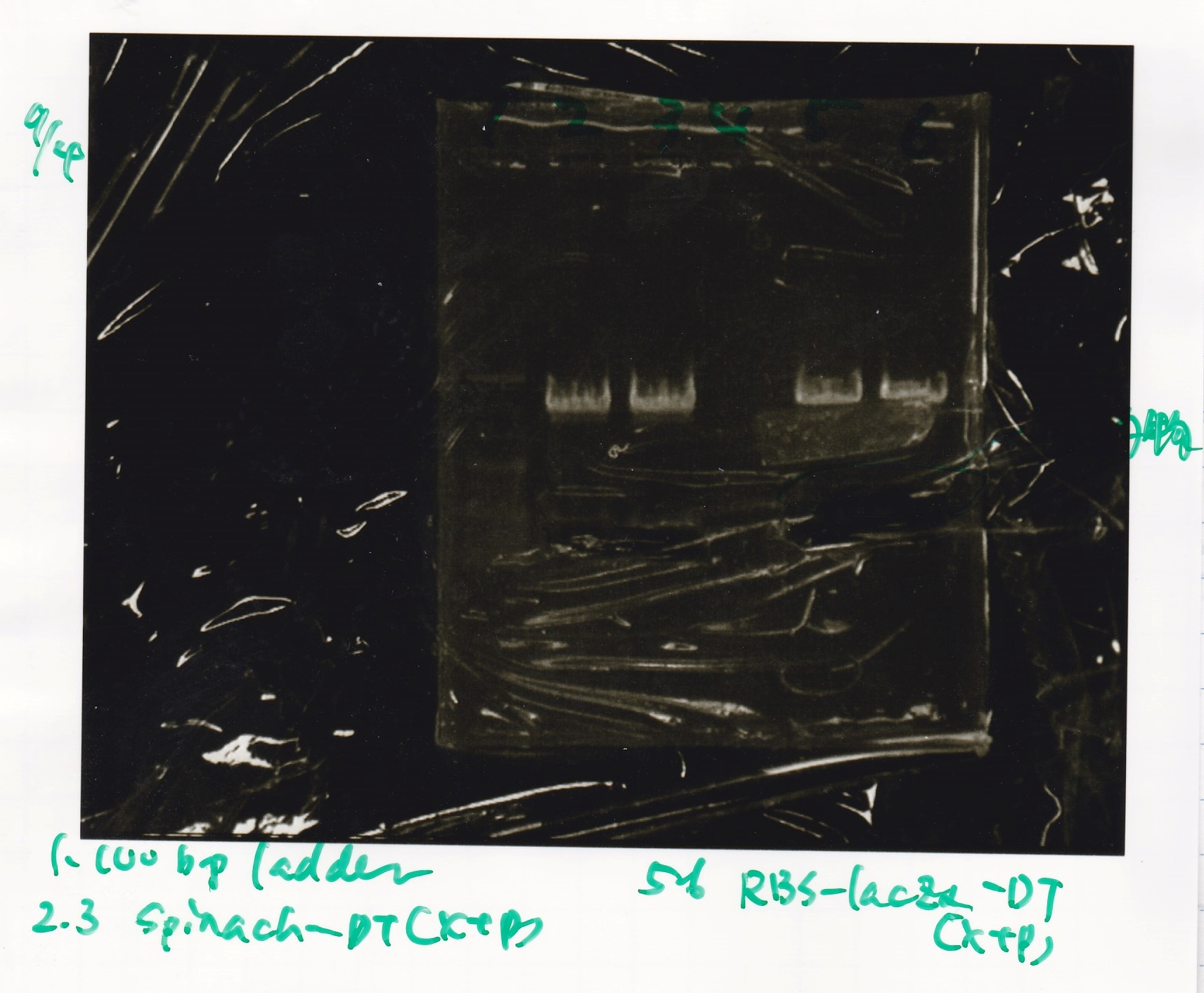
| Name | quantity | concentration[µg/mL] | 260/280 | 260/230
|
| spinach-DT | -- | 2.1 | 2.10 | -0.05
|
| RBS-lacZα-DT | -- | 3.3 | 2.59 | -0.07
|
Master Plate
No name
| Number | Use LB plate (+Amp)
|
| 4 | 8/9 BBa_J23100-3
|
| 5 | 8/9 BBa_J23100-4
|
| 6 | 8/9 BBa_J23100-5
|
| Number | Use LB plate
|
| 1 | JM109-1
|
| 2 | JM109-2
|
| 3 | JM109-3
|
| 4 | JM109-4
|
Liquid Culture
Hirano
| Sample | medium
|
| 8/9 BBa_J23100-3 | Plusgrow medium(+Amp)
|
| JM109-1(Master plate) | LB medium
|
Colony PCR
Hirano
| Sample | base pair
|
| 8/9 BBa_J23100 3 | --
|
| 8/9 BBa_J23100 4 | --
|
| 8/9 BBa_J23100 5 | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
Sep 5
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | J23100-3 | -- | --
|
| 3 | J23100-4 | -- | --
|
| 4 | J23100-5 | -- | --
|
| 5 | 100bp ladder | -- | --
|
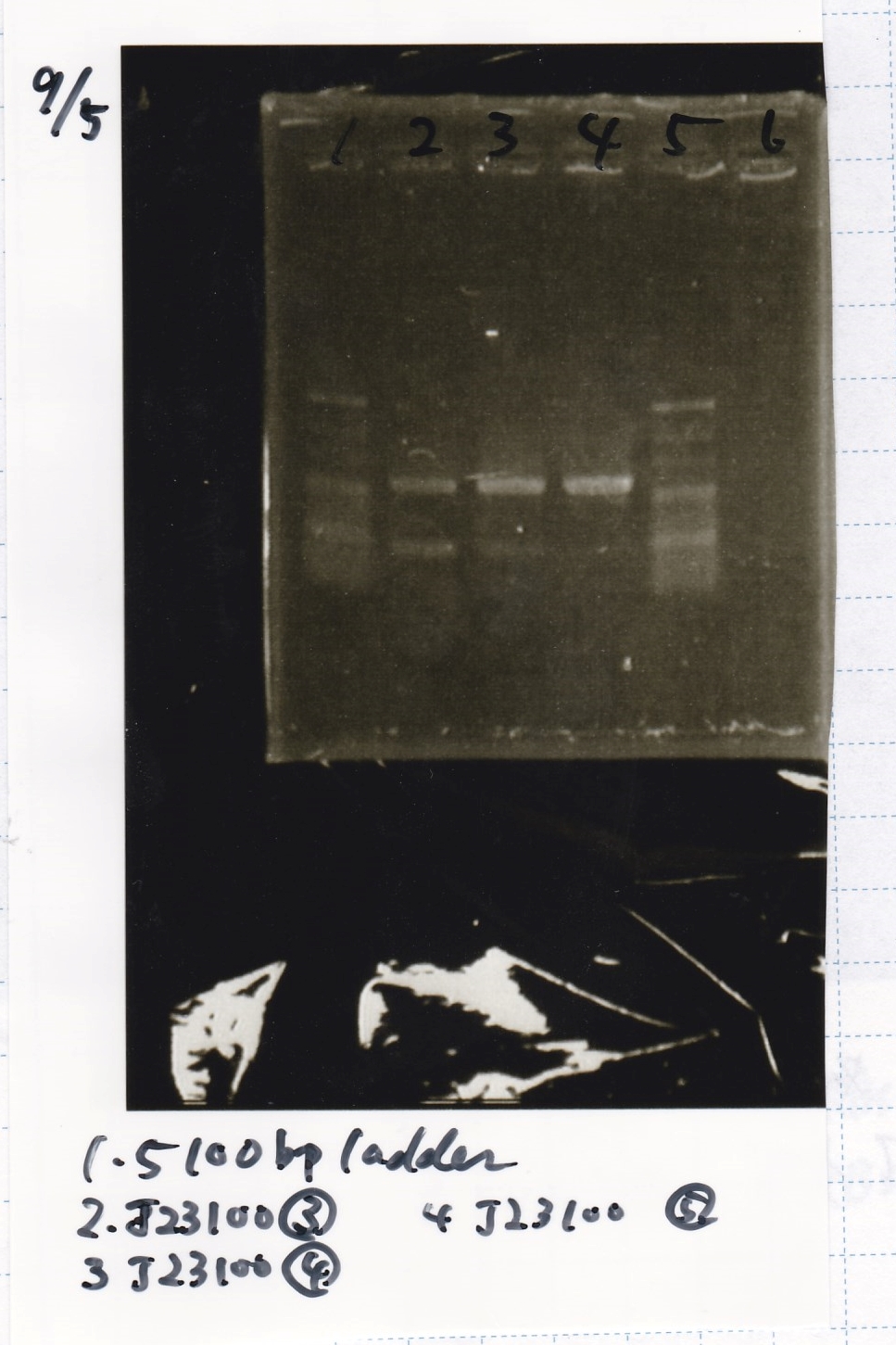
Miniprep
Nakamoto
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 8/3 J23100-3 | 349.1 | 1.69 | 1.851
|
| 8/10 Plac-2 | 331.3 | 1.84 | 1.42
|
Ligation
Kojima
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | Pconst | 13.2 | spinach-DT | 2.1 | 3.5
|
| experiment | Plac | 15.3 | spinach-DT | 2.1 | 3.5
|
| experiment | Pconst | 13.2 | pT181 antisense (XbaI & PstI) | 6.6 | 3.5
|
| experiment | Plac | 15.3 | pT181 antisense (XbaI & PstI) | 6.6 | 3.5
|
| experiment | Ptet | 30.1 | pT181 antisense (XbaI & PstI) | 6.6 | 3.5
|
| experiment | Pconst | 13.2 | pT181 attenuator | 11.7 | 3.5
|
| experiment | Plac | 15.3 | pT181 attenuator | 11.7 | 3.5
|
| experiment | Ptet | 30.1 | RBS-lacZα-DT | 3.0 | 3.5
|
incubate 16 °C 1 hour
Electrophoresis
Tatsui
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | RBS-luxI-DT
|
| 3 | Pbad/araC
|
| 4 | NC
|
| 5 | 100bp ladder
|

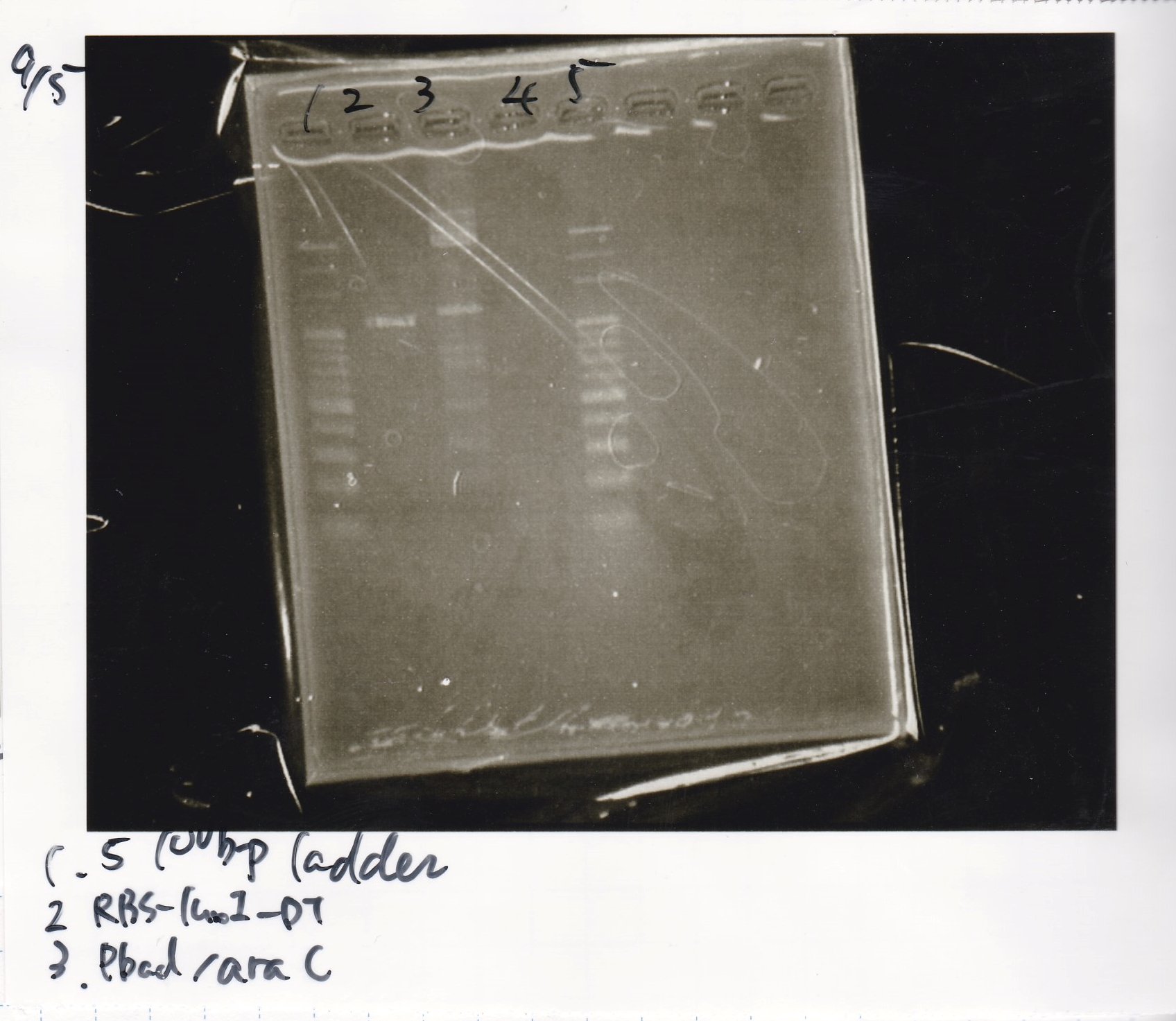
No name
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder |
|
| 2 | RBS-luxI-DT |
|
| 3 | RBS-luxI-DT |
|
| 4 | |
|
| 5 | Pbad/araC |
|
| 6 | Pbad/araC |
|


| Name | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-luxI-DT | 23.2 | 0.83 | 7.89
|
| Pbad/araC | 51.8 | 1.20 | 0.11
|
Colony PCR
No name
| Sample | base pair
|
| 9/4 aptamer 12_1R(pSB1C3)-1 | 384
|
| 9/4 aptamer 12_1R(pSB1C3)-2 | 384
|
| 9/4 pT181 antisense(pSB1C3)-1 | 415
|
| 9/4 pT181 antisense(pSB1C3)-2 | 415
|
| 9/4 pT181 antisense(pSB1C3)-3 | 415
|
| 9/4 pT181 antisense(pSB1C3)-4 | 415
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 30s | 30cycles
|
| Sample | base pair
|
| 9/4 aptamer 12_1R-DT-1 | 521
|
| 9/4 aptamer 12_1R-DT-2 | 521
|
| 9/4 aptamer 12_1R-DT-3 | 521
|
| 9/4 aptamer 12_1R-DT-4 | 521
|
| 9/4 pT181 attenuator(pSB1C3) | 601
|
| 9/4 pT181 attenuator(pSB1C3) | 601
|
| 9/4 RBS-lysis1-DT | 613
|
| 9/4 RBS-lysis1-DT | 613
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 36s | 30cycles
|
| Sample | base pair
|
| 9/4 RBS-lysis2-DT-1 | 985
|
| 9/4 RBS-lysis2-DT-2 | 985
|
| 9/4 RBS-lysis2-DT-3 | 985
|
| 9/4 RBS-lysis2-DT-4 | 985
|
| 9/4 RBS-lysis3-DT-1 | 1210
|
| 9/4 RBS-lysis3-DT-2 | 1210
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 72s | 30cycles
|
Liquid Culture
Nakamoto
| Sample | medium
|
| 8/9 J23100-5 | Plusgrow medium(+Amp)
|
37°C
Transformation
No name
| Name | Sample(µL) | Competent Cells(µL) | Total(µL) | Plate
|
| 9/5 Pconst+spinach-DT | 2 | 20 | 22 | --
|
| 9/5 Plac+spinach-DT | 2 | 20 | 22 | --
|
| 9/5 Pconst+pT181 antisense | 2 | 20 | 22 | --
|
| 9/5 Plac+pT181 antisense | 2 | 20 | 22 | --
|
| 9/5 Ptet+pT181 antisense | 2 | 20 | 22 | --
|
| 9/5 Plux+pT181 attenuator | 2 | 20 | 22 | --
|
| 9/5 Ptet+pT181 attenuator | 2 | 20 | 22 | --
|
| 9/5 Ptet+pT181 attenuator | 2 | 20 | 22 | --
|
| 9/5 Pconst+RBS-tetR+DT | 2 | 20 | 22 | --
|
| 9/16(2012) T7-His-FT | 2 | 20 | 22 | --
|
| 9/16(2012) pBr322 | 2 | 20 | 22 | --
|
Restriction Enzyme Digestion
No name
| | 9/5 Pbad/araC | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 4µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 18µL | 30µL
|
| | 9/5 RBS-luxI-DT | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 4µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 18µL | 30µL
|
| | 8/28 Plux | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 12µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 10µL | 30µL
|
| NC | 0.6µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.4µL | 10µL
|
| | 9/5 Pconst(J23100) | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 5.7µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 16.3µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 9/5 Plac | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 6.0µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 16µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 8/31 Plux-RBS-GFP-DT-1 | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 9.4µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 12.6µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| | 8/20 Pconst-RBS-luxR-DT | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 4.5µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 13.5µL | 30µL
|
| NC | 0.2µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.8µL | 10µL
|
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/5 RBS-luxI-DT | 7.6 | 1.56 | -1.42
|
| 9/5 Pbad/araC | 7.5 | 1.54 | 3.84
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | aptamer 12_1R (pSB1C3)-1 | -- | --
|
| 3 | aptamer 12_1R (pSB1C3)-2 | -- | --
|
| 4 | pT181 antisense(pSB1C3)-1 | -- | --
|
| 5 | pT181 antisense(pSB1C3)-2 | -- | --
|
| 6 | pT181 antisense(pSB1C3)-3 | -- | --
|
| 7 | pT181 antisense(pSB1C3)-4 | -- | --
|
| 8 | 100bp ladder | -- | --
|
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | aptamer 12_1R-DT-1 | -- | --
|
| 3 | aptamer 12_1R-DT-2 | -- | --
|
| 4 | aptamer 12_1R-DT-3 | -- | --
|
| 5 | aptamer 12_1R-DT-4 | -- | --
|
| 6 | pT181 attenuator(pSB1C3)-1 | -- | --
|
| 7 | pT181 attenuator(pSB1C3)-2 | -- | --
|
| 8 | RBS-lysis1-DT-1 | -- | --
|
| 9 | RBS-lysis1-DT-2 | -- | --
|
| 10 | 100bp ladder | -- | --
|
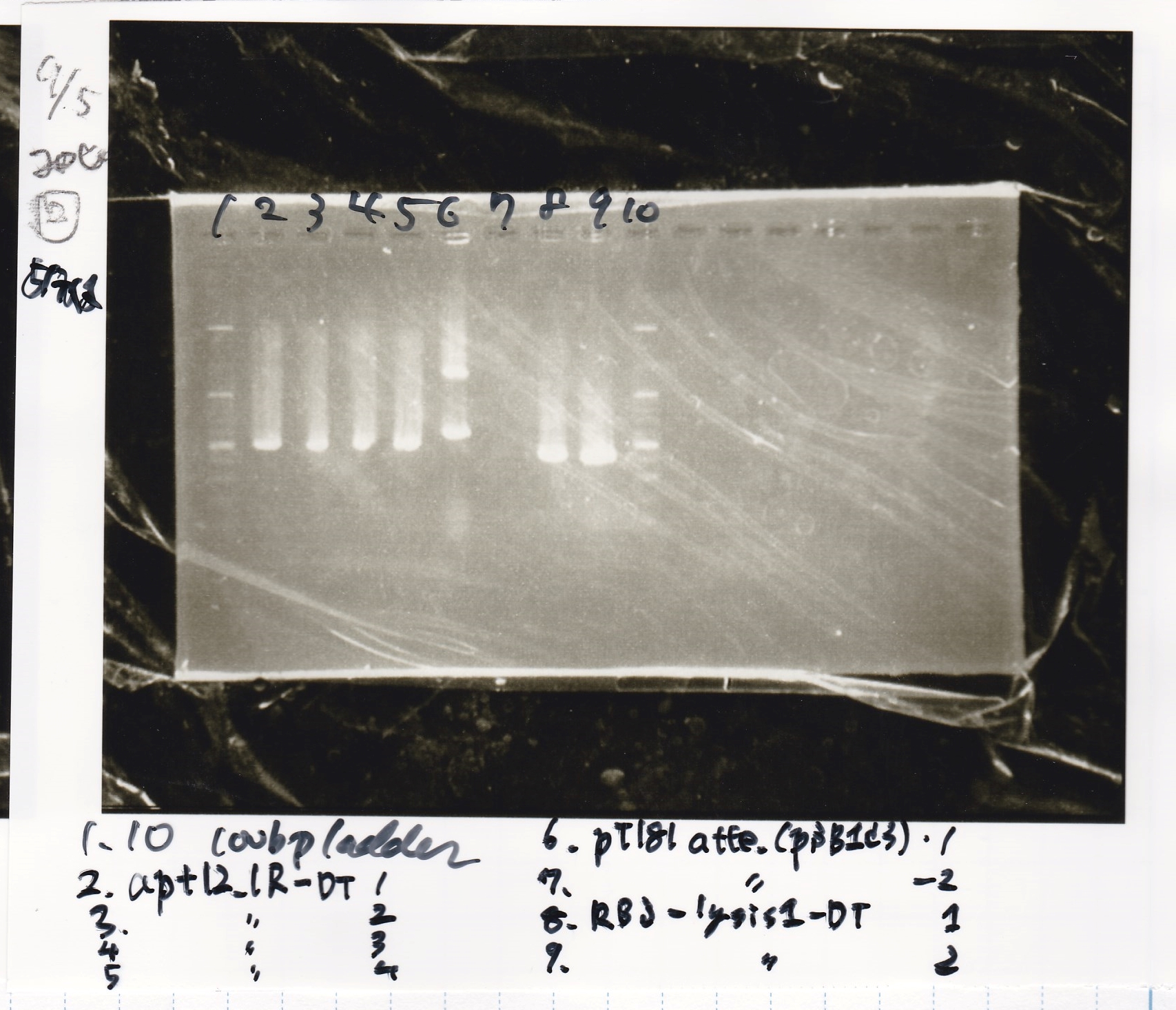
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | RBS-lysis2-DT-1 | -- | --
|
| 3 | RBS-lysis2-DT-2 | -- | --
|
| 4 | RBS-lysis2-DT-3 | -- | --
|
| 5 | RBS-lysis2-DT-4 | -- | --
|
| 6 | RBS-lysis3-DT-1 | -- | --
|
| 7 | RBS-lysis3-DT-2 | -- | --
|
| 8 | 100bp ladder | -- | --
|
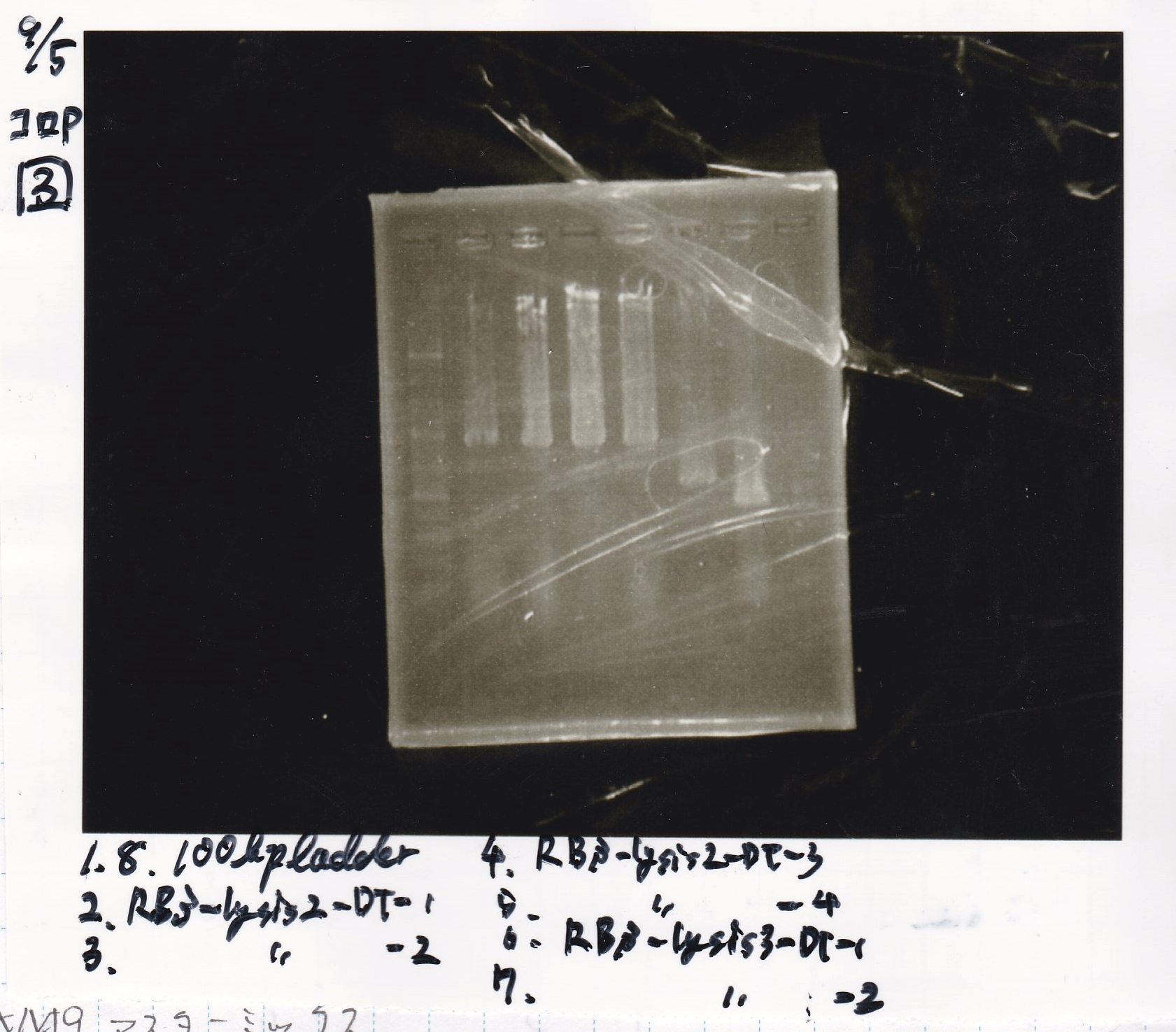
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kbp ladder | -- | --
|
| 2 | Pconst | SpeI | PstI
|
| 3 | Pconst NC | -- | --
|
| 4 | Plac | SpeI | PstI
|
| 5 | Plac NC | -- | --
|
| 6 | Plux | SpeI | PstI
|
| 7 | Plux NC | -- | --
|
| 8 | Pconst-RBS-luxR-DT | EcoRI | XbaI
|
| 9 | Pconst-RBS-luxR-DT NC | -- | --
|
| 10 | Plux-RBS-GFP-DT | EcoRI | SpeI
|
| 11 | Plux-RBS-GFP-DT NC | -- | --
|
| 12 | 1kbp ladder | -- | --
|
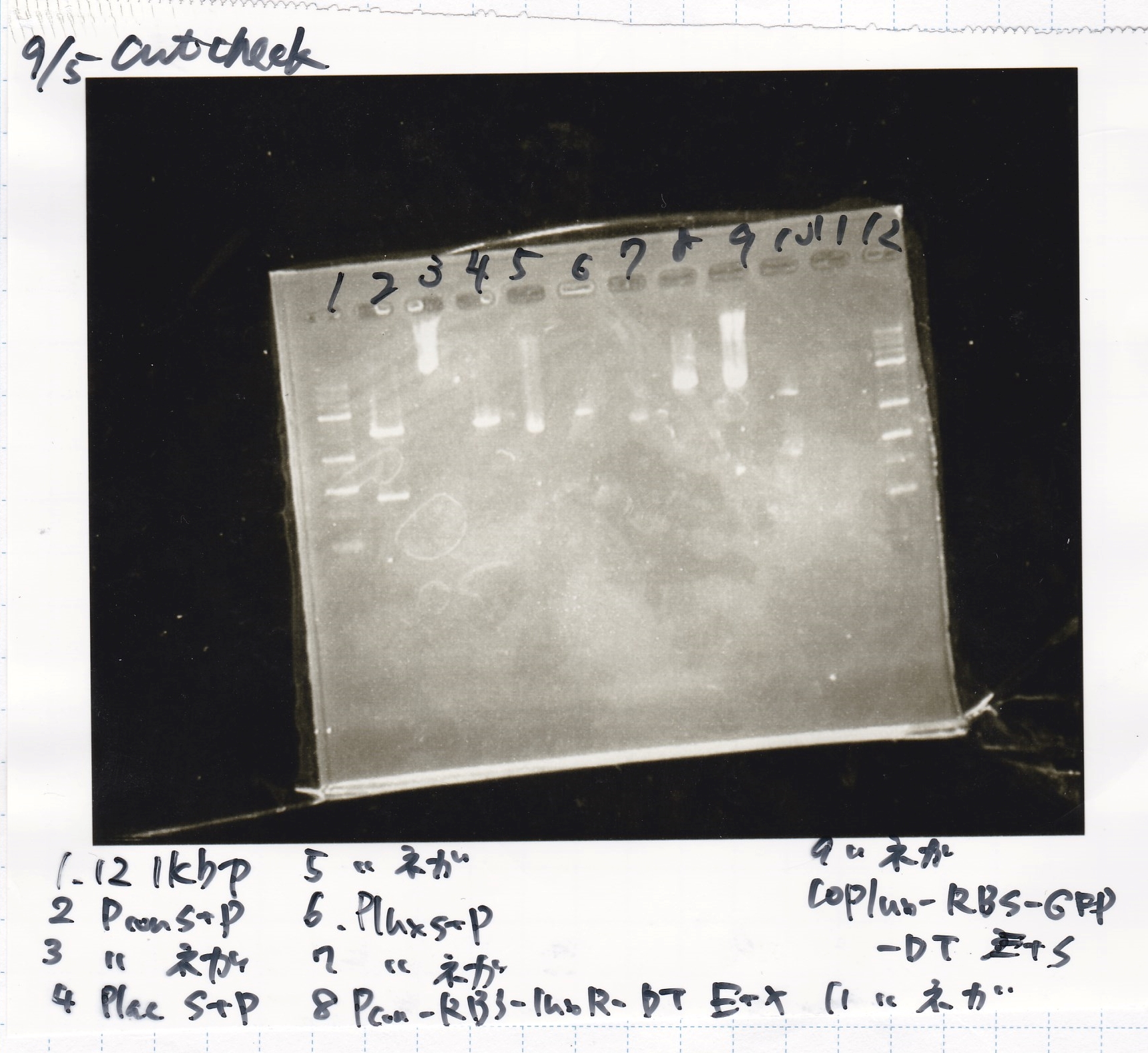
5x M9 Medium (+EDTA)
Hirano
| volume | 10ml
|
| Na2HPO4 | 60mg
|
| KH2PO4 | 30mg
|
| NaCl | 5mg
|
| NH4Cl | 10mg
|
| Fe(III)-EDTA | 1263.27mg
|
| MilliQ | up to 10 mL
|
- autoclave at 121 °C for 20 min
No name
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder |
|
| 2 | Pconst | SpeI & PstI
|
| 3 | Pconst | SpeI & PstI
|
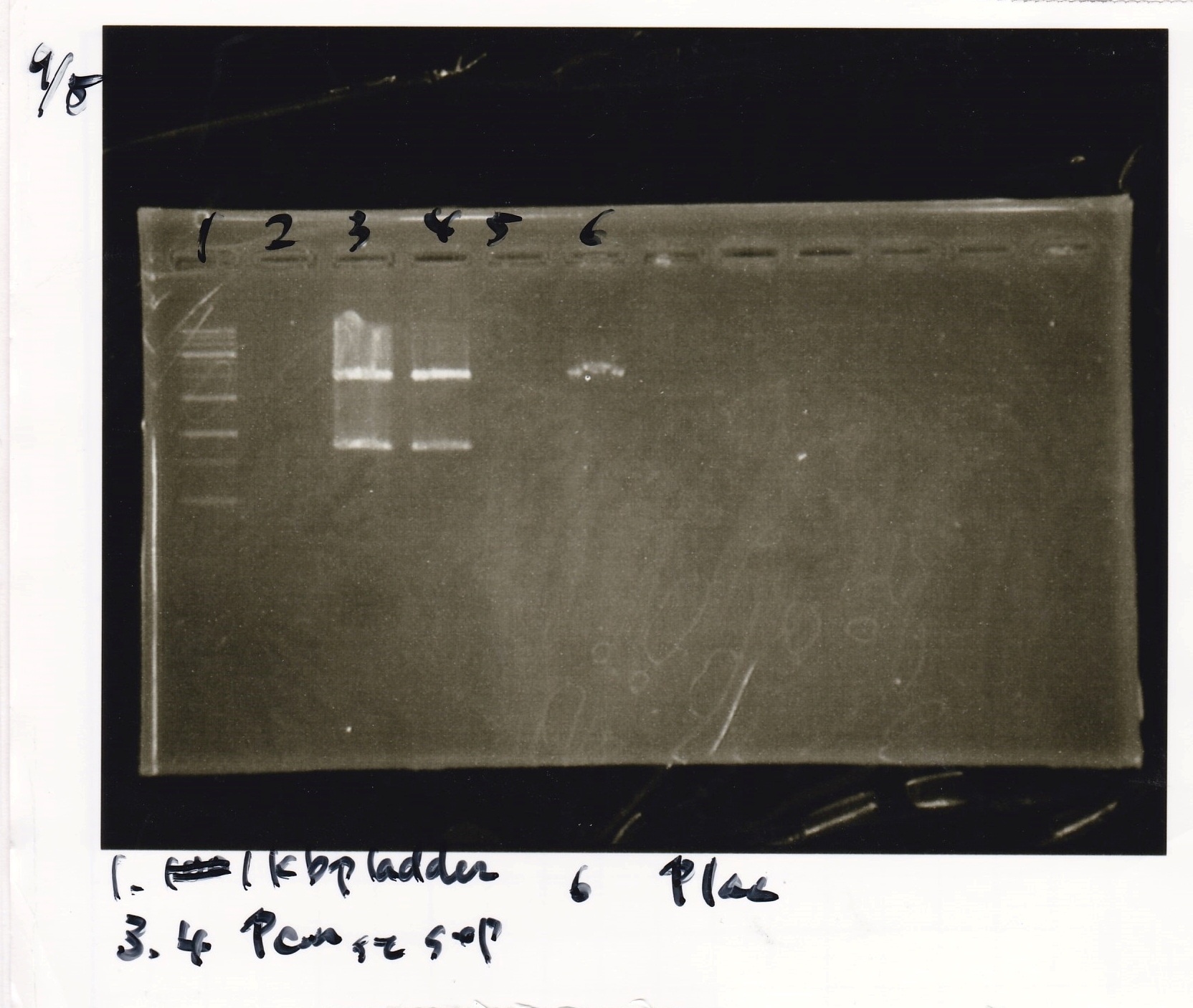

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pconst | 5.3 | 1.98 | 0.06
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 2 | Plac | SpeI & PstI
|
| 3 | Plac | SpeI & PstI
|
| 5 | Plux | SpeI & PstI
|
| 6 | Plux | SpeI & PstI
|
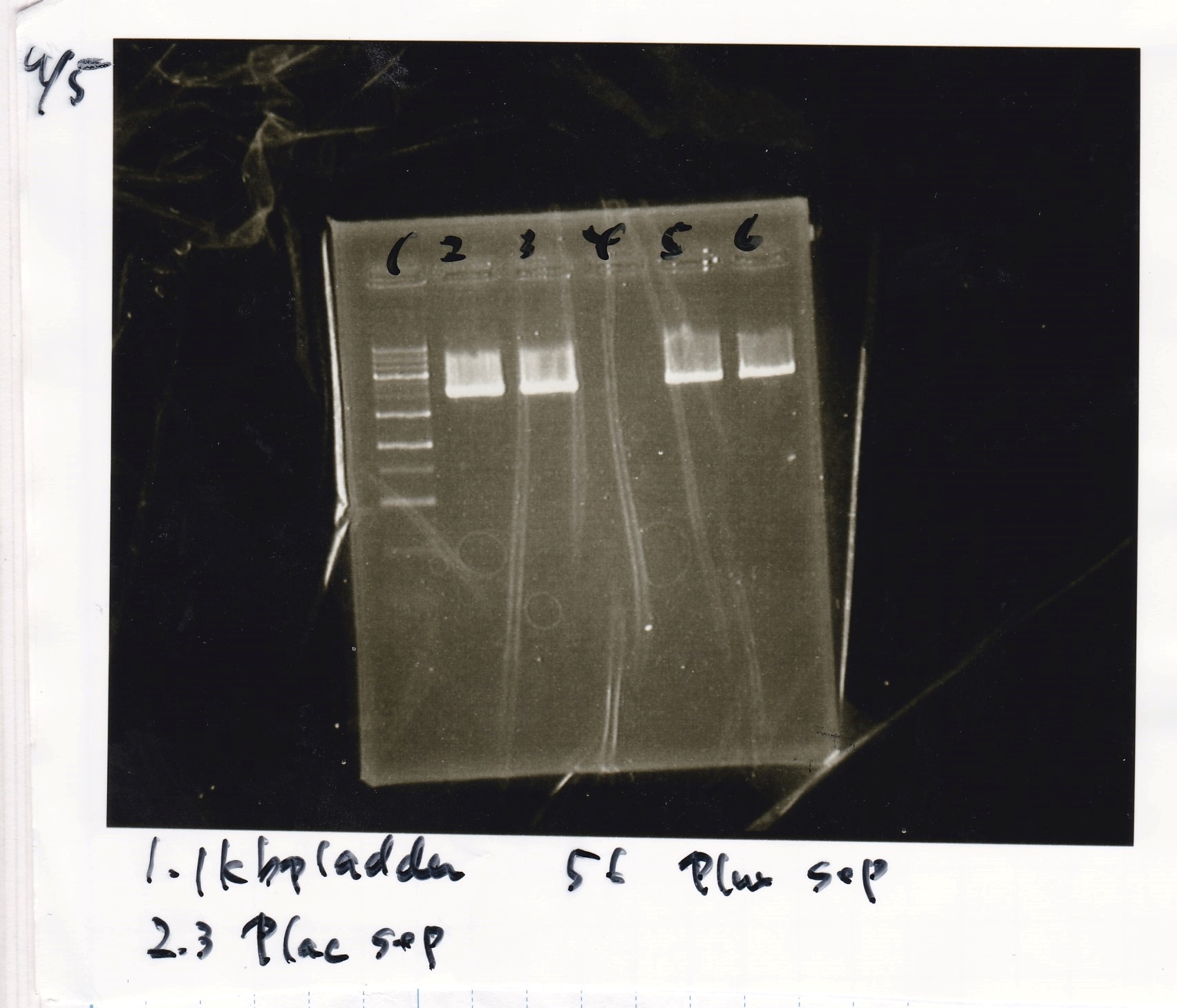

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Plac (SpeI & PstI) | 5.2 | 1.82 | 0.36
|
| Plux(SpeI & PstI) | 8.0 | 1.87 | 0.25
|
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder |
|
| 2 | Pconst-RBS-luxR-DT | EcoRI & XbaI
|
| 3 | Pconst-RBS-luxR-DT | EcoRI & XbaI
|
| 5 | Plux-RBS-GFP-DT | EcoRI & SpeI
|
| 6 | Plux-RBS-GFP-DT | EcoRI & SpeI
|
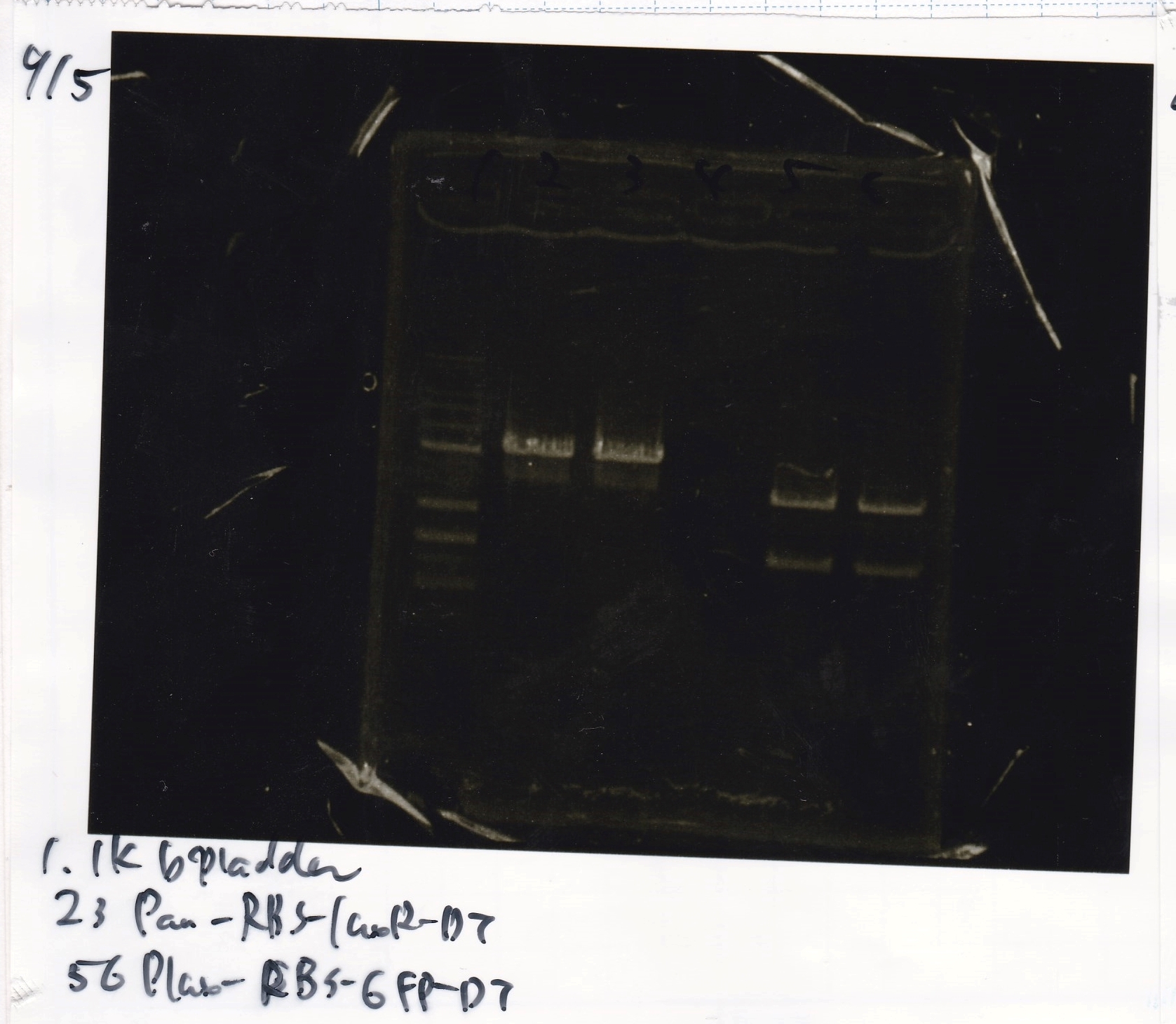

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pconst-RBS-luxR-DT(EcoRI & XbaI) | 30.6 | 1.84 | 1.16
|
| Plux-RBS-GFP-DT (EcoRI & SpeI) | 21.8 | 1.87 | 0.98
|
DNA Purification
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/5 RBS-luxI-DT | 7.6 | 1.56 | -1.42
|
| 9/5 Pbad/araC | 7.5 | 1.54 | 3.84
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 8/9 J23100-5 | 243.0 | 1.90 | 1.81
|
Colony PCR
No name
| Sample | base pair
|
| 9/4 RBS-lysis3-DT-3 | 1210
|
| 9/4 RBS-lysis3-DT-4 | 1210
|
| 9/4 RBS-lysis3-DT-5 | 1210
|
| 9/4 RBS-lysis1-DT-3 | 613
|
| 9/4 Ptrc KaiC-1 | --
|
| 9/4 pSB4K5 | 1370
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min 25s | 30cycles
|
Liquid Culture
Hirano
| Sample | medium
|
| 9/4 aptamer 12_1R-DT-1 | Plusgrow medium (+CP)
|
| 9/4 pT181 attenuator(pSB1C3)-1 | Plusgrow medium (+CP)
|
| 9/4 RBS-lysis2-DT-1 | Plusgrow medium (+CP)
|
| 9/4 Ptrc-KaiC -1 | Plusgrow medium (+Amp)
|
| 9/4 pSB4K5 -1 | Plusgrow medium (+Kan)
|
Sep 6
Electrophoresis
Hirano
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | 9/4 RBS-lysis3-DT -3
|
| 3 | 9/4 RBS-lysis3-DT -4
|
| 4 | 9/4 RBS-lysis3-DT -5
|
| 5 | 9/4 RBS-lysis1-DT -3
|
| 6 | 9/4 Ptrc-KaiC -1
|
| 7 | 9/4 pSB4K5 -1
|
| 8 | 100bp ladder
|
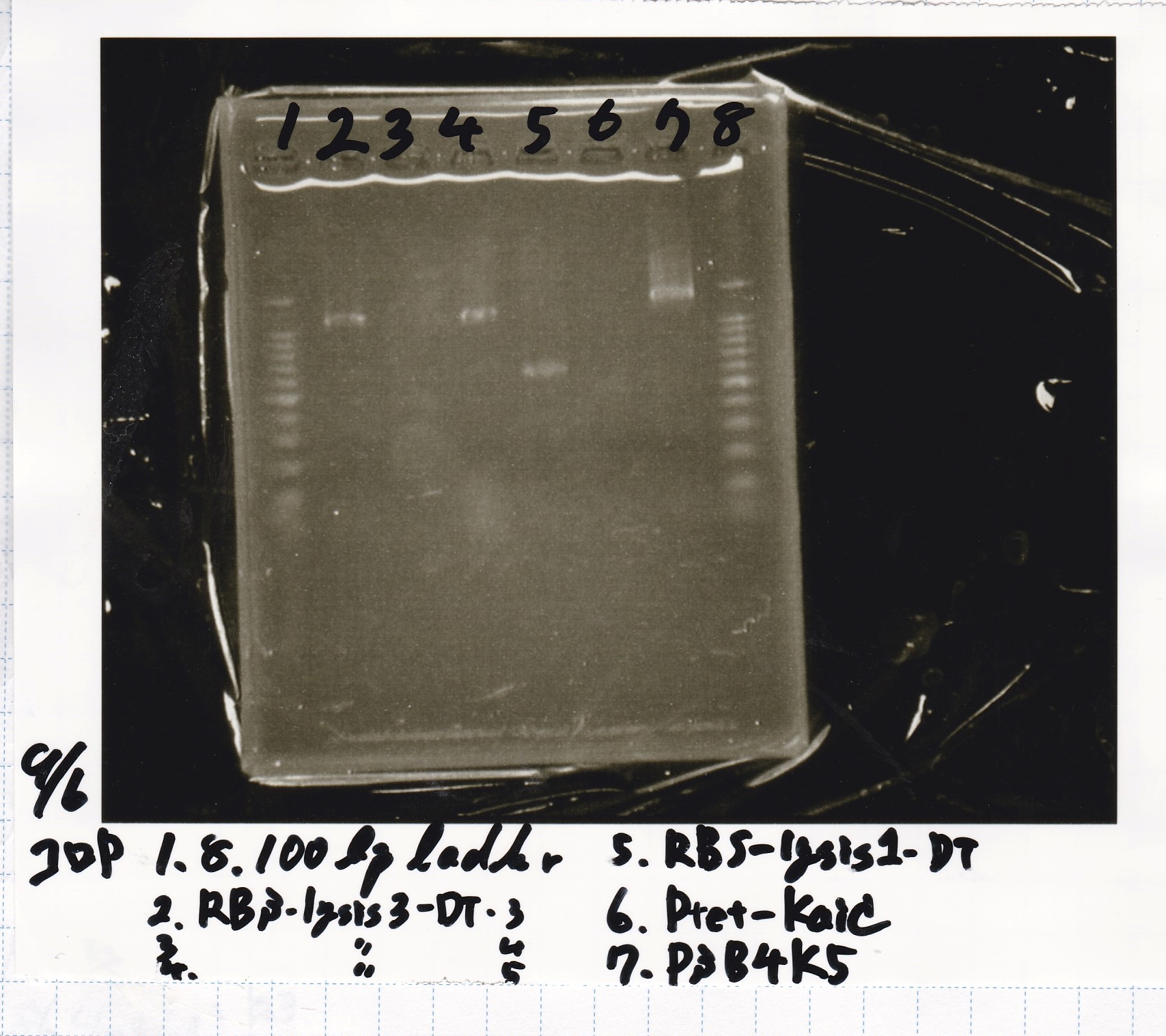
Colony PCR
No name
| Sample | base pair
|
| Pconst-spinach-DT-(1~4) | 563
|
| Plac-spinach-DT-(1~4) | 659
|
| Pcon-pT181 antisense-(1~4) | 468
|
| Ptet-pT181 antisense-(1~4) | 604
|
| Pconst-pT181 attenuator-(1~4) | 568
|
| Pconst+RBS-tetR-DT-(1~4) | 1121
|
| Ptet+RBS-lacZα-DT-(1~4) | 1162
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 40s | 30cycles
|
Liquid Culture
No name
| Sample | medium
|
| 9/4 RBS-Lysis3+DT3 | Plusgrow medium(+CP)
|
| 9/4 RBS-Lysis1+DT3 | Plusgrow medium(+CP)
|
| 9/4 pSB4K5 | Plusgrow medium(+Kan)
|
Electrophoresis
No name
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Pconst-spinach-DT -(1)
|
| 3 | Pconst-spinach-DT -(2)
|
| 4 | Pconst-spinach-DT -(3)
|
| 5 | Pconst-spinach-DT -(4)
|
| 6 | Plac-spinach-DT -(1)
|
| 7 | Plac-spinach-DT -(2)
|
| 8 | Plac-spinach-DT -(3)
|
| 9 | Plac-spinach-DT -(4)
|
| 10 | Pcon-pT181 antisense -(1)
|
| 11 | Pcon-pT181 antisense -(2)
|
| 12 | Pcon-pT181 antisense -(3)
|
| 13 | Pcon-pT181 antisense -(4)
|
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Ptet-pT181 antisense -(1)
|
| 3 | Ptet-pT181 antisense -(2)
|
| 4 | Ptet-pT181 antisense -(3)
|
| 5 | Ptet-pT181 antisense -(4)
|
| 6 | Plac-pT181 attenuator -(1)
|
| 7 | Plac-pT181 attenuator -(2)
|
| 8 | Plac-pT181 attenuator -(3)
|
| 9 | Plac-pT181 attenuator -(4)
|

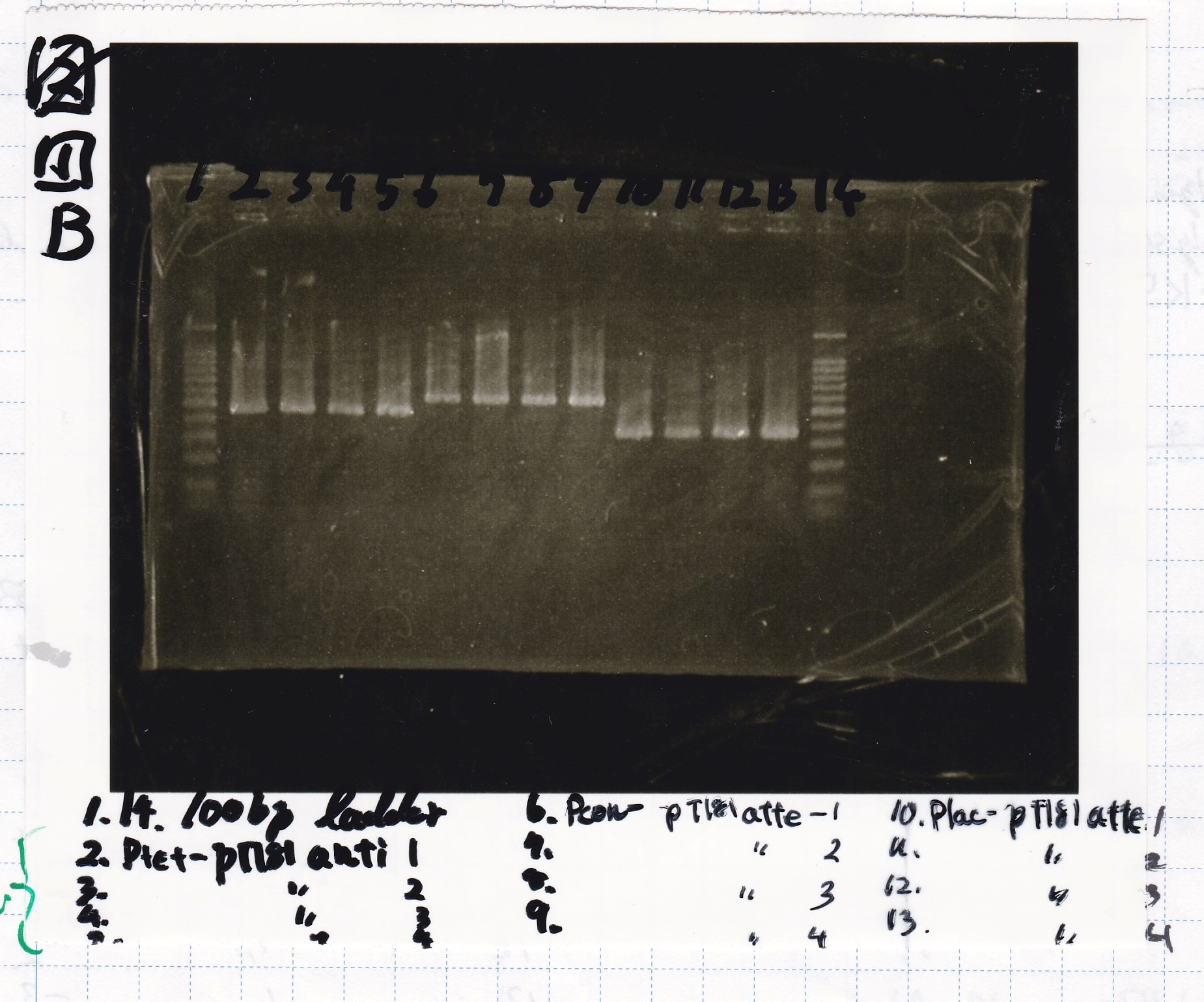
Miniprep
Honda and Kojima
| DNA | concentration [µg/mL] | 260/280 | 260/230
|
| pT181 antisense-(1) | 238.7 | 1.68 | 1.44
|
| pT181 antisense-(2) | 248.8 | 1.83 | 1.77
|
| pT181 antisense-(3) | 223.2 | 1.78 | 1.61
|
| pT181 antisense-(4) | 220.8 | 1.89 | 1.92
|
| tetR aptamer-(1) | 252.5 | 1.89 | 1.90
|
| tetR aptamer-(2) | 211.2 | 1.90 | 1.99
|
| DT181 attenuator | 102.2 | 1.98 | 2.17
|
| 121R aptamer-DT | 251.7 | 1.92 | 2.01
|
| pSB4K5 | 212.9 | 1.80 | 1.90
|
| RBS-lacZα-DT | 185.7 | 1.78 | 1.79
|
| RBS-lysis2-DT-(1) | 84.8 | 1.85 | 2.24
|
Restriction Enzyme Digestion
Tatsui
| | 9/6 RBS-lysis2-DT (78.3 µg/mL) | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 22 µL | 1 µL | 1 µL | 3 µL | 3 µL | 0 µL | 30 µL
|
| NC | 1.3 µL | 0 µL | 0 µL | 1 µL | 1 µL | 6.7 µL | 10 µL
|
| | aptamer-DT (251.7 µg/mL) | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 0.4 µL | 1 µL | 1 µL | 3 µL | 3 µL | 14.1 µL | 30 µL
|
| NC | 0.4 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.6 µL | 10 µL
|
| | Pλ-luxI (86.4 µg/mL) | EcoRI | SpeI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 23.1 µL | 1 µL | 1 µL | 3 µL | 3 µL | 1.9 µL | 30 µL
|
| NC | 1.2 µL | 0 µL | 0 µL | 1 µL | 1 µL | 6.8 µL | 10 µL
|
Electrophoresis
Hirano
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Pconst+RBS-tetR-DT -(1)
|
| 3 | Pconst+RBS-tetR-DT -(2)
|
| 4 | Pconst+RBS-tetR-DT -(3)
|
| 5 | Pconst+RBS-tetR-DT -(4)
|
| 6 | Ptet+RBS-lacZα-DT -(1)
|
| 7 | Ptet+RBS-lacZα-DT -(2)
|
| 8 | 100bp ladder
|
| 9 | Ptet+RBS-lacZα-DT -(3)
|
| 10 | Ptet+RBS-lacZα-DT -(4)
|
| 11 | 100bp ladder
|

No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | RBS-lysis2-DT | XbaI & PstI
|
| 3
|
| 4 | -- | --
|
| 5 | aptamer 12_1R-DT | XbaI & PstI
|
| 6
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | Pλ-luxI | EcoRI & SpeI
|
| 3
|
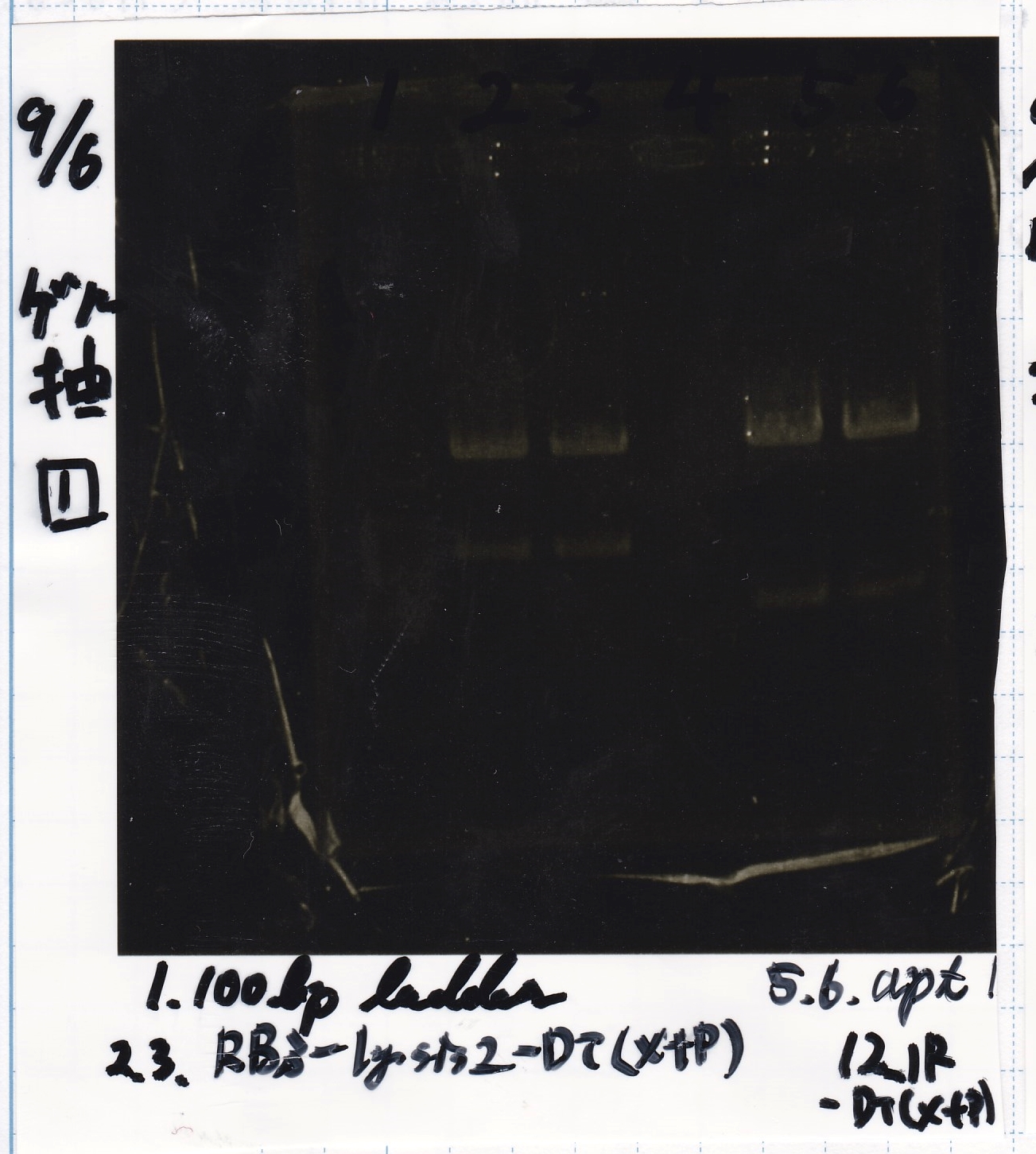

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-lysis2-DT (XbaI & PstI) | 7.1 | 1.88 | 0.07
|
| aptamer 12_1R-DT (XbaI & PstI) | 15.3 | 1.93 | 0.70
|
| Pλ-luxI (EcoRI & SpeI) | 4.5 | 1.79 | 0.31
|
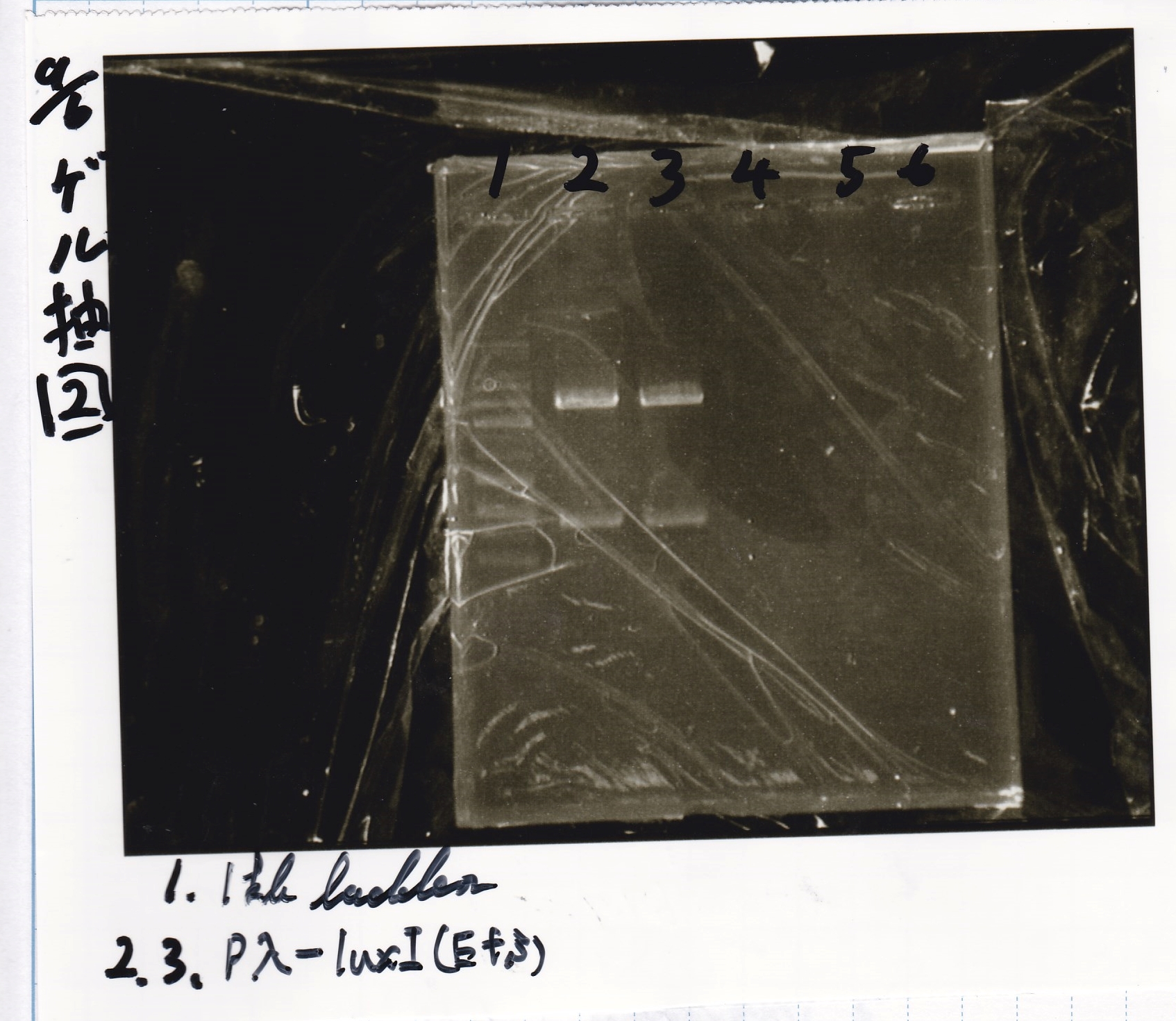

Liquid Culture
No name
| Sample | medium
|
| 9/5 Pconst-spinach-DT-(1) | Plusgrow medium(+Amp&CP)
|
| 9/5 Ptet-pT181 antisense-(1) | Plusgrow medium(+CP)
|
| 9/5 Pconst+RBS-tetR-DT-(1) | Plusgrow medium(+Amp&CP)
|
| 9/5 Ptet+RBS-lacZα-DT-(1) | Plusgrow medium(+CP)
|
| 9/5 Pcon-pT181 attenuator-(1) | Plusgrow medium(+CP)
|
| 9/4 pT181 antisense-(1) | Plusgrow medium(+CP)
|
| 9/4 aptamer 12_1R-(1) | Plusgrow medium(+CP)
|
Colony PCR
No name
| Sample | base pair
|
| 9/5 Plac-spinach-DT-(5~12) | 563
|
| 9/5 Pcon-antisense-(5~12) | 468
|
| 9/5 Plac-attenuator-(5~12) | 664
|
| 9/5 pT181 antisense+Plac-(1) | -
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 42s | 30cycles
|
Sep 7
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-lysis1-DT | 195.3 | |
|
| RBS-lysis3-DT | 255.5 | 1.91 | 2.04
|
| Ptet-pT18 | 183.6 | 1.85 | 1.48
|
| Pcon-RBS-tetR-DT | 134.3 | 1.94 | 2.20
|
| Ptet-RBS-lacZα-DT | 67.7 | 1.92 | 1.79
|
| Pcon-pT181 attenuator | 232.3 | 1.95 | 1.90
|
| pSB4K5 | 237.0 | 1.61 | 1.50
|
| pT181 antisense(pSB1C3) | 256.4 | 1.63 | 1.78
|
| apt12_1R(pSB1C3) | 230.0 | 1.98 | 2.28
|
Electrophoresis
No name
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Plac-Spinach-DT -5
|
| 3 | Plac-Spinach-DT -6
|
| 4 | Plac-Spinach-DT -7
|
| 5 | Plac-Spinach-DT -8
|
| 6 | Plac-Spinach-DT -9
|
| 7 | Plac-Spinach-DT -10
|
| 8 | 100bp ladder
|
| 1 | 100bp ladder
|
| 2 | Plac-Spinach-DT -11
|
| 3 | Plac-Spinach-DT -12
|
| 4 | Pcon-antisense -5
|
| 5 | Pcon-antisense -6
|
| 6 | Pcon-antisense -7
|
| 7 | Pcon-antisense -8
|
| 8 | 100bp ladder
|

| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Pcon-antisense -9
|
| 3 | Pcon-antisense -10
|
| 4 | Pcon-antisense -11
|
| 5 | Pcon-antisense -12
|
| 6 | Plac-attenuator -5
|
| 7 | Plac-attenuator -6
|
| 8 | 100bp ladder
|
| 1 | 100bp ladder
|
| 2 | Plac-attenuator -7
|
| 3 | Plac-attenuator -8
|
| 4 | Plac-attenuator -9
|
| 5 | Plac-attenuator -10
|
| 6 | Plac-attenuator -11
|
| 7 | Plac-attenuator -12
|
| 8 | 100bp ladder
|

| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Plac-pT181 antisense
|

Colony PCR
No name
| Sample
|
| Plac-spinach-DT -13~20
|
| Plac-pT181 attenuator -13~20
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 94 °C | 55 °C | 68°C | --
|
| 5 min | 30 s | 30 s | 42 sec | 30 cycles
|
Ligation
No Name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/6 Plux (SpeI&PstI) | 2 µL | 9/7 RBS-lysis2-DT (EcoRI&XbaI) | µL | 3.5 µL
|
| experiment | 9/6 Plac (SpeI&PstI) | µL | 9/7 RBS-lysis2-DT (EcoRI&XbaI) | µL | 3.5 µL
|
| experiment | 9/6 Pcon-RBS-luxR-DT (EcoRI&XbaI) | 1.9 µL | 9/6 Plux-RBS-GFP-DT(SpeI&PstI) | 2.2 µL | 3.5 µL
|
| experiment | 9/6 Pbed/araC (SpeI&PstI) | 1.6 µL | 9/5 RBS-luxI-DT (EcoRI&XbaI) | 16 µL | 3.5 µL
|
| experiment | 9/6 Plac (SpeI&PstI) | 1.5 µL | 9/5 pT181 antisense (EcoRI&XbaI) | 2.0 µL | 3.5 µL
|
| experiment | 9/6 Pcon (SpeI&PstI) | 3.0 µL | 9/5 RBS-luxI-DT (EcoRI&XbaI) | 10 µL | 3.5 µL
|
Electrophoresis
Nakamoto
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Plac-Spinach-DT -13
|
| 3 | Plac-Spinach-DT -14
|
| 4 | Plac-Spinach-DT -15
|
| 5 | Plac-Spinach-DT -16
|
| 6 | Plac-Spinach-DT -17
|
| 7 | Plac-Spinach-DT -18
|
| 8 | Plac-Spinach-DT -19
|
| 9 | Plac-Spinach-DT -20
|
| 10 | Plac-pT181 attenuator -13
|
| 11 | Plac-pT181 attenuator -14
|
| 12 | 100bp ladder
|
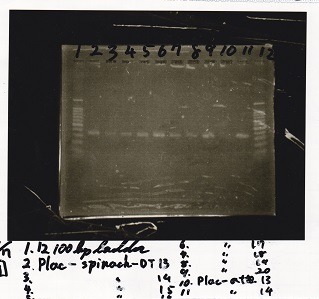
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Plac-pT181 attenuator -15
|
| 3 | Plac-pT181 attenuator -16
|
| 4 | Plac-pT181 attenuator -17
|
| 5 | Plac-pT181 attenuator -18
|
| 6 | Plac-pT181 attenuator -19
|
| 7 | Plac-pT181 attenuator -20
|
| 8 | 100bp ladder
|
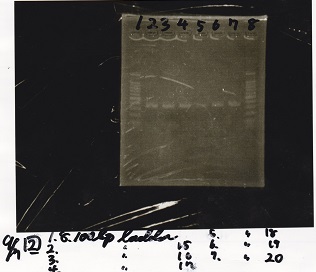
Transformation
Tatsui
| Name | Sample | Competent Cells | Total | Plate
|
| Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT | 2µL | 20µL | 22µL |
|
| Pbed/araC+RBS-luxI-DT | 2µL | 20µL | 22µL |
|
| Plac+pT181 antisense | 2µL | 20µL | 22µL |
|
| Pcon+RBS-luxI-DT | 2µL | 20µL | 22µL |
|
Restriction Enzyme Digestion
No Name
| | DNA | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 9/7 RBS-lysis1-DT(XbaI & PstI) | 10 µL | 1 µL | 1 µL | 3 µL | 3 µL | 12 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.5 µL | 10 µL
|
| | DNA | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 9/7 RBS-lysis3-DT (XbaI & PstI) | 7.8 µL | 1 µL | 1 µL | 3 µL | 3 µL | 14.2 µL | 30 µL
|
| NC | 0.4 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.6 µL | 10 µL
|
| | DNA | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 9/5 Plac (SpeI & PstI) | 6.0 µL | 1 µL | 1 µL | 3 µL | 3 µL | 16.0 µL | 30 µL
|
| NC | 0.3 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.7 µL | 10 µL
|
| | DNA | EcorI | SpeI | Buffer | BSA | MilliQ | total
|
| 9/7 pSB4K5 (EcoRI & SpeI) | 8.4 µL | 1 µL | 1 µL | 3 µL | 3 µL | 16.0 µL | 30 µL
|
| NC | 0.4 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.7 µL | 10 µL
|
| | DNA | restricion enzyme | restriction enzyme | Buffer | BSA | MilliQ | total
|
| Pcon-pT181 attenuator (SpeI & PstI) | 8.6 µL | 1 µL | 1 µL | 3 µL | 3 µL | 13.4 µL | 30 µL
|
| Pcon-pT181 attenuator (EcorI & SpeI) | 8.6 µL | 1 µL | 1 µL | 3 µL | 3 µL | 13.4 µL | 30 µL
|
| NC | 0.4 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.6 µL | 10 µL
|
| | DNA | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 9/5 Spinach-DT (XbaI & PstI) | 5.4 µL | 1 µL | 1 µL | 3 µL | 3 µL | 16.6 µL | 30 µL
|
| NC | 0.3 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.7 µL | 10 µL
|
| | DNA | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| Ptet-anticodone (SpeI & PstI) | 11 µL | 1 µL | 1 µL | 3 µL | 3 µL | 11 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.5 µL | 10 µL
|
| | DNA | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 9/6 RBS-lacZα-DT (XbaI & PstI) | 11 µL | 1 µL | 1 µL | 3 µL | 3 µL | 11 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.5 µL | 10 µL
|
| | DNA | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| pSB1C3 (1) (EcoRI & SpeI) | 6.6 µL | 1 µL | 1 µL | 3 µL | 3 µL | 15.4 µL | 30 µL
|
| NC | 0.3 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.7 µL | 10 µL
|
| | DNA | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| pSB1C3 (3) (XbaI & PstI) | 6.6 µL | 1 µL | 1 µL | 3 µL | 3 µL | 15.4 µL | 30 µL
|
| NC | 0.3 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.7 µL | 10 µL
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | RBS-lysis1-DT (9/7) | XbaI | PstI
|
| 3 | RBS-lysis1-DT (9/7) | -- | --
|
| 4 | RBS-lysis3-DT (9/7) | XbaI | PstI
|
| 5 | RBS-lysis3-DT (9/7) | -- | --
|
| 6 | Pcon-pT181 attenuator | EcoRI | SpeI
|
| 7 | Pcon-pT181 attenuator | -- | --
|
| 8 | 9/5 Spinach-DT | XbaI | PstI
|
| 9 | 9/5 Spinach-DT | -- | --
|
| 10 | 100bp ladder | -- | --
|
| 11 | 100bp ladder | -- | --
|
| 12 | RBS-lacZα-DT | XbaI | PstI
|
| 13 | RBS-lacZα-DT | -- | --
|
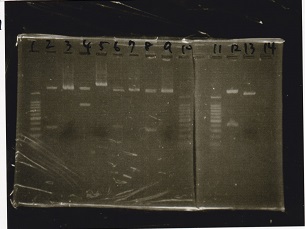
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Ptet-antisense | SpeI | PstI
|
| 3 | Plac (NC) | -- | --
|
| 4 | pSB4K5 | EcoRI | SpeI
|
| 5 | pSB4K5 | -- | --
|
| 6 | Pcon-pT181 attenuator | SpeI | PstI
|
| 7 | 1kbp ladder | -- | --
|

| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Plac | SpeI | PstI
|
| 3 | Ptet-antisense | -- | --
|
| 4 | pSB1C3 | EcoRI | SpeI
|
| 5 | pSB1C3 | -- | --
|
| 6 | pSB1C3 | XbaI | PstI
|
| 7 | pSB1C3 | -- | --
|
| 8 | 1kbp ladder | -- | --
|

Liquid Culture
No name
| Sample | medium
|
| entA- strain | SOB medium (+Kan)
|
| 9/15 Pcon-pT181 antisense | Plusgrow (+Amp)
|
Sep 8
No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 2 | RBS-lysis1-DT | XbaI & PstI
|
| 3 | RBS-lysis1-DT | XbaI & PstI
|
| 5 | RBS-lysis3-DT | XbaI & PstI
|
| 6 | RBS-lysis3-DT | XbaI & PstI
|
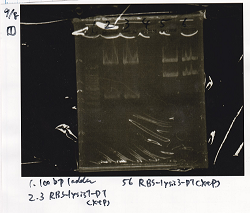

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-lysis1-DT (XbaI & PstI) | 5.3 | 1.98 | 0.06
|
| RBS-lysis3-DT (XbaI & PstI) | 5.3 | 1.98 | 0.06
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 3 | Pcon-pT181 attenuator | EcoRI & SpeI
|
| 4 | Pcon-pT181 attenuator | EcoRI & SpeI
|
| 6 | Spinach-DT | XbaI & PstI
|
| 7 | Spinach-DT | XbaI & PstI
|
| 9 | RBS-lacZα-DT | XbaI & PstI
|
| 10 | RBS-lacZα-DT | XbaI & PstI
|

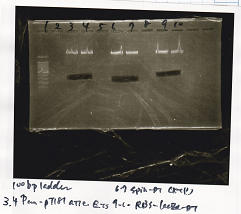
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-pT181 attenuator (EcoRI & SpeI) | 5.2 | 1.82 | 0.36
|
| Spinach-DT (XbaI & PstI) | 8.0 | 1.87 | 0.25
|
| RBS-lacZα-DT (XbaI & PstI) | 10.3 | 1.71 | 0.54
|
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder |
|
| 2 | Ptet-pT181 antisense | EcoRI & PstI
|
| 3 | Ptet-pT181 antisense | EcoRI & PstI
|
| 5 | pSB4K5 | EcoRI & SpeI
|
| 6 | pSB4K5 | EcoRI & SpeI
|


| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Ptet-pT181 antisense(EcoRI & PstI) | 30.6 | 1.84 | 1.16
|
| pSB4K5 (EcoRI & SpeI) | 21.8 | 1.87 | 0.98
|
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder |
|
| 2 | Pcon-pT181 attenuator | SpeI & PstI
|
| 3 | Pcon-pT181 attenuator | SpeI & PstI
|
| 5 | Plac | SpeI & PstI
|
| 6 | Plac | SpeI & PstI
|
| 8 | pSB1C3 | EcoRI & SpeI
|
| 9 | pSB1C3 | EcoRI & SpeI
|
| 11 | pSB1C3 | XbaI & PstI
|
| 12 | pSB1C3 | XbaI & PstI
|


| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-pT181 attenuator (SpeI & PstI) | 14.5 | 1.91 | 1.09
|
| Plac (SpeI & PstI) | 11.6 | 1.74 | 1.19
|
| pSB1C3 (EcoRI & SpeI) | 5.7 | 1.74 | 0.34
|
| pSB1C3 (XbaI & PstI) | 5.6 | 2.00 | 0.37
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-pT181 anti | 243.0 | 1.90 | 1.81
|
| Pcon-Spinach-DT | 253.7 | 1.57 | 1.51
|
Ligation
Kojima
| state | Vector(µL) | Inserter(µL) | Ligation High ver.2
|
| experiment | 9/8 Plac SpeI+PstI | 4.3 | 9/3 pT181 attenuator | 3.1 | 3.5
|
| experiment | 9/8 Plac SpeI+PstI | 4.3 | 9/7 aptamer12-1R-DT XbaI+PstI | 2.1 | 3.2
|
| experiment | 9/8 Plac SpeI+PstI | 4.3 | 9/8 spinach-DT XbaI+PstI | 4.6 | 3.5
|
| experiment | 9/6 Plac SpeI+PstI | 2 | 9/7 RBS-lysis2-DT XbaI+PstI | 10 | 3.5
|
| experiment | 9/8 Plac SpeI+PstI | 4.3 | 9/7 RBS-lysis2-DT XbaI+PstI | 10 | 3.5
|
| experiment | 9/8 Ptet-pT181 anti | 1.6 | 9/8 spinach-DT XbaI+PstI | 4.3 | 3.0
|
| experiment | 9/6 Pcon SpeI+PstI | 1.4 | 9/8 RBS-laczα-DT XbaI+PstI | 4.8 | 3.1
|
| experiment | 9/8 Pcon-pT181 attenuator SpeI+PstI | 3.4 | 9/7 aptamer12-1R-DT XbaI+PstI | 1.9 | 2.7
|
| experiment | 9/6 Pcon SpeI+PstI | 1.4 | 9/7 aptamer12-1R-DT XbaI+PstI | 2.1 | 1.3
|
| experiment | 9/8 DT | 3.0 | 9/8 Pcon-pT181 attenuator EcoRI+SpeI | 7.9 | 3.5
|
| experiment | 9/6 Plux SpeI+PstI | 2 | 9/7 RBS-lysis1-DT XbaI+PstI | 7.2 | 3.5
|
| experiment | 9/8 Plac SpeI+PstI | 4.3 | 9/7 RBS-lysis1-DT XbaI+PstI | 7.2 | 3.5
|
| experiment | 9/4 Plac SpeI+PstI | 1.7 | 9/8 RBS-laczα-DT XbaI+PstI | 4.8 | 3.3
|
Restriction Enzyme Digestion
Kojima
| | 9/8 Pcon-pT181 anti | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 8.2µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 13.8µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 9/8 Pcon-spinach-DT | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 7.9µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 14.1µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 8/21 F1 attenuator | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 6.5µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 15.5µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 8/21 F3m2 attenuator | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 6.0µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 16µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 8/21 F1 anti | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 6.7µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 15.3µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 8/21 F6 anti | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 6.0µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 16µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 8/21 tetR aptamer 12_1M | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 5.3µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 16.7µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 8/21 tetR aptamer 12_P | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 13.3µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 8.7µL | 30µL
|
| NC | 0.7µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL
|
| | 9/4 spinach-DT | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 5.4µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 16.6µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 9/7 RBS-lysis1-DT | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 10µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 12µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| | 9/6 RBS-lysis2-DT | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 13µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 9µL | 30µL
|
| NC | 0.7µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL
|
| | 9/7 RBS-lysis3-DT | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 10µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 12µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| | 8/20 J23100 RBS-lacZα-DT-(1) | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 7µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 3µL | 16µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| | 8/20 RBS-lacZα-DT | EcoRI | SpeI | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 10µL | 0.5µL | 0.5µL | 0µL | 0µL | 3µL | 3µL | 13µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
Transformation
Nakamoto & Ashida
| Name | Sample(µL) | Competent Cells(µL) | Total(µL) | Plate
|
| 9/8 Pcont+aptamer 12-1R-DT | 1 | 10 | 11 | Amp
|
| 9/8 Pcon+RBS-lacZα-DT | 1 | 10 | 11 | Amp
|
| 9/8 Pcon-pT181 attenuator+aptamer 12-1R-DT | 1 | 10 | 11 | Amp
|
| 9/8 Plac+RBS-lysis1-DT | 2 | 20 | 22 | CP
|
| 9/8 Plac+RBS-lysis2-DT | 2 | 20 | 22 | CP
|
| 9/8 Plac+aptamer 12-1R-DT | 2 | 20 | 22 | CP
|
| 9/8 Plac+spinach | 2 | 20 | 22 | CP
|
| 9/8 Plac+pT181 attenuator | 2 | 20 | 22 | CP
|
| 9/8 Plux+RBS-lysis1-DT | 1 | 10 | 11 | CP
|
| 9/8 Plux+RBS-lysis2-DT | 1 | 10 | 11 | CP
|
| 9/8 Ptet+RBS-lacZα-DT | 1 | 10 | 11 | CP
|
| 9/8 Ptet-pT181 anti+spinach-DT | 1 | 10 | 11 | CP
|
| 9/8 Pcon-pT181 attenuator+DT | 1 | 10 | 11 | CP
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | |
|
| 2 | Pcon-spinach-DT | EcoRI | SpeI
|
| 3 | Pcon-spinach-DT | |
|
| 4 | Fusion1 attenuator | XbaI | PstI
|
| 5 | Fusion1 attenuator | |
|
| 6 | Fusion3m2 attenuator | XbaI | PstI
|
| 7 | Fusion3m2 attenuator | |
|
| 8 | Fusion1 anti | XbaI | PstI
|
| 9 | Fusion1 anti | |
|
| 10 | Fusion6 anti | XbaI | PstI
|
| 11 | Fusion6 anti | |
|
| 12 | aptamer 12-1M | XbaI | PstI
|
| 13 | aptamer 12-1M | |
|
| 14 | aptamer 12-P | XbaI | PstI
|
| 15 | aptamer 12-P | |
|
| 16 | spinach-DT | XbaI | PstI
|
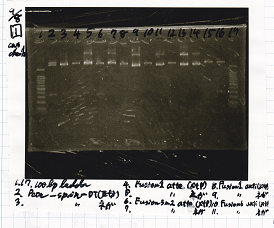
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | spinach-DT | |
|
| 2 | J23100-RBS-lacZα-DT | EcoRI | SpeI
|
| 3 | J23100-RBS-lacZα-DT | |
|
| 4 | 100bp ladder | |
|
| 5 | RBS-lacZα-DT | EcoRI | SpeI
|
| 6 | RBS-lacZα-DT | |
|
| 7 | RBS-lysis1-DT | XbaI | PstI
|
| 8 | RBS-lysis1-DT | |
|
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | |
|
| 2 | RBS-lysis2-DT | XbaI | PstI
|
| 3 | RBS-lysis2-DT | |
|
| 4 | RBS-lysis3-DT | XbaI | PstI
|
| 5 | RBS-lysis3-DT | |
|
| 6 | Pcon pT181 anti | SpeI | PstI
|
| 7 | Pcon pT181 anti | |
|

Ligation
No name
| state | Vector | Inserter
|
| experiment | Pcon-spinach-DT(EcoRI&SpeI) | 10.5µg/mL | pSB6A1(EcoRI&SpeI) | 8.9µg/mL
|
| experiment | Pcon-spinach-DT(EcoRI&SpeI) | 10.5µg/mL | pSB4K5(EcoRI&SpeI) | 21.8µg/mL
|
| experiment | Pcon-pT181 anti(SpeI+PstI) | 31.3µg/mL | spinach-DT(XbaI+PstI) | 10.1µg/mL
|
| experiment | Plac(SpeI+PstI) | 11.6µg/mL | RBS-lysis3-DT | 23.4µg/mL
|
| experiment | Plux(SpeI+PstI) | 25µg/mL | RBS-lysis3-DT | 23.4µg/mL
|
| experiment | Ptet-pT181 anti(SpeI+PstI) | 30.6µg/mL | spinach-DT(XbaI+PstI) | 10.1µg/mL
|
| experiment | Pcon-pT181 attenuator(SpeI+PstI) | 14.5µg/mL | RBS-laczα-DT(XbaI+PstI) | 4.4µg/mL
|
PCR
No name
| KaiA(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(661-RBS-KaiA-fwd) | Primer(kaiA-662-rev-ver2) | MilliQ | total
|
| 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 28s | 30cycles
|
| KaiB(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(662-RBS-KaiB-fwd-ver2) | Primer(kaiB-663-rev-ver2) | MilliQ | total
|
| 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 28s | 30cycles
|
| DT(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(664-DT-fwd-ver2) | Primer(DT-suffix-660-rev-ver2) | MilliQ | total
|
| 0.1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.4 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 28s | 30cycles
|
| P-KaiB(PCR prpduct) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(prefix-PkaiBC-fwd-ver2) | Primer(PkaiBC-suffix--662-rev-ver2) | MilliQ | total
|
| 0.7 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.8 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 28s | 30cycles
|
| Plac(1)(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(GG0-pSBIC3-fwd-ver2) | Primer(Plac-GG1-rev-ver2) | MilliQ | total
|
| 0.13 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.37 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 60s | 30cycles
|
| KaiC(plasmid) | KOD plus | 10x buffer | dNTP | MgSO4 | Primer(GG3-RBS-KaiC-fwd-ver2) | Primer(KaiC-GG4-rev-ver2) | MilliQ | total
|
| 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 60s | 30cycles
|
Yoshida
| Lane | DNA
|
| 1 | 100bp ladder
|
| 2 | KaiA
|
| 3 | KaiA
|
| 4 | KaiB
|
| 5 | KaiB
|
| 6 | DT
|
| 7 | DT
|
| 8 | P-KaiBC
|
| 9 | P-KaiBC
|
| 10 | nega
|
| 11 | 100bp ladder
|
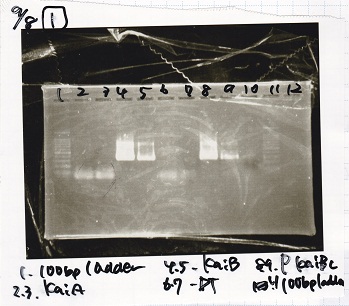

| Lane | DNA
|
| 1 | Plac
|
| 2 | Plac
|
| 3 | KaiC
|
| 4 | KaiC
|
| 5 | nega
|
| 6 | 1kb ladder
|

Sep 9
PCR
Nakagawa
| KaiA(plasmid) | 10x buffer | dNTPs | MgSO4 | Primer(661-RBS-KaiA-fwd) | Primer(kaiA-662-rev-ver2) | Kod-plus ver2 | MilliQ | total
|
| 1.0 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 58.5°C | 68°C | --
|
| 2min | 10s | 30s | 30s | 30cycles
|
| KaiB(plasmid) | 10x buffer | dNTPs | MgSO4 | Primer(661-RBS-KaiA-fwd-ver2) | Primer(kaiB-663-rev-ver2) | Kod-plus ver2 | MilliQ | total
|
| 1.0 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 58.5°C | 68°C | --
|
| 2min | 10s | 30s | 30s | 30cycles
|
| P-KaiBC(PCR products) | 10x buffer | dNTPs | MgSO4 | Primer(prefix-P-kaiBC-fwd-ver2) | Primer(P-KaiBC-suffix-rev-verw) | Kod-plus ver2 | MilliQ | total
|
| 0.7 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 15.8 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 58.5°C | 68°C | --
|
| 2min | 10s | 30s | 30s | 30cycles
|
| DT(plasmid) | 10x buffer | dNTPs | MgSO4 | Primer(664-DT-fwd-ver2) | Primer(DT-suffix-660-rev-ver2) | Kod-plus ver2 | MilliQ | total
|
| 1.0 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 0.5 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 58.5°C | 68°C | --
|
| 2min | 10s | 30s | 30s | 30cycles
|
Yoshida
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | KaiA | --
|
| 3
|
| 4 | KaiB | --
|
| 5
|
| 6 | P-KaiBC | --
|
| 7
|
| 8 | DT | --
|
| 9
|
| 10 | NC | --
|
| 11 | 100bp ladder | --
|

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| KaiC | 28.5 | 1.26 | 11.92
|
| Plac | 53.9 | 1.82 | 1.61
|
| KaiB | 59.6 | 1.61 | 2.20
|
| KaiB | 76.5 | 1.86 | 1.44
|
| DT | 98.7 | 1.47 | 1.43
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/8 pSB1C3 | 83.2 | 1.45 | 0.76
|
| 9/8 RBS-lysis2-DT | 72.8 | 1.67 | 0.97
|
| 9/8 spinach-DT | 152.6 | 1.88 | 1.97
|
| Nuclease-Free water | -0.4 | -14.30 | 0.25
|
Restriction Enzyme Digestion
No name
| | 9/9 pSB1C3 | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 24µL | 1µL | 1µL | 3µL | 3µL | 22µL | 30µL*
|
| NC | 1.2µL | 0µL | 0µL | 1µL | 1µL | 6.8 µL | 10µL
|
| | 9/9 pSB1C3 | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 27.4µL | 1µL | 1µL | 3µL | 3µL | 22µL | 30µL*
|
| NC | 1.3µL | 0µL | 0µL | 1µL | 1µL | 6.7µL | 10µL
|
| | 9/9 spinach-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 10.8µL | 1µL | 1µL | 3µL | 3µL | 22µL | 30µL*
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
Transformation
No name
| Name | Sample | Competent Cells | Total | Plate
|
| Pcon+RBS-luxI-DT | 2µL | 20µL | 22µL | Amp
|
| Pcon-RBS-luxR-DT+Plux-GFP | 2µL | 20µL | 22µL |
|
| Ptet-PT181 antisense+spinach-DT-Pbad/araC+RBS-lux-DT | 2µL | 20µL | 22µL |
|
Ligation
No Name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/8 F1-attenuator(XbaI&PstI) | 5.3µg | 9/8 pSB1C3(XbaI&PstI) | 8.9µg | 3.5µg
|
| experiment | 9/8 F3m2-attenuator(XbaI&PstI) | 4.5µg | 9/8 pSB1C3(XbaI&PstI) | 8.9µg | 3.5µg
|
| experiment | 9/8 F1-antisense(XbaI&PstI) | 1.7µg | 9/8 pSB1C3(XbaI&PstI) | 8.9µg | 3.5µg
|
| experiment | 9/8 F6-antisense(XbaI&PstI) | 2.9µg | 9/8 pSB1C3(XbaI&PstI) | 8.9µg | 3.5µg
|
| experiment | 9/8 aptamer12-P(EcoRI&SpeI) | 3.1µg | 9/8 pSB1C3(EcoRI&SpeI) | 8.8µg | 3.5µg
|
| experiment | 9/8 aptamer12-1M(EcoRI&SpeI) | 3.1µg | 9/8 pSB1C(EcoRI&SpeI)3 | 8.8µg | 3.5µg
|
M9 medium
Shimizu & Hirano
| EDTA consenlation | Na2HPO4 | KH2PO4 | NaCl | NH4Cl | Fe(Ⅲ)-EDTA | distilled water | total
|
| 1mMol | 300mg | 150mg | 25mg | 50mg | 21mg | up to 10mL | 10mL
|
| 10mMol | 300mg | 150mg | 25mg | 50mg | 210.5mg | up to 10mL | 10mL
|
| 50mMol | 300mg | 150mg | 25mg | 50mg | 1052.7mg | up to 10mL | 10mL
|
Colony PCR
No name
| Sample | base pair
|
| 9/8 Plac-spinach-DT-(1) | 659
|
| 9/8 Plac-spinach-DT-(2) | 659
|
| 9/8 Pcon-pt181attenuator-DT-(1) | 584
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 40s | 30cycles
|
No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | Spinach-DT | XbaI+PstI
|
| 3
|
| 5 | RBS-lacZα-DT | XbaI+PstI
|
| 6
|

| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 3 | pSB1C3 | XbaI+PstI
|
| 4
|
Ligation
No name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | pSB1C3(XbaI+PstI) | xx µL | 9/8 F1-attenuator(XbaI+PstI) | xx µL | xx µL
|
| experiment | pSB1C3(XbaI+PstI) | xx µL | 9/8 F3m2-attenuator(XbaI+PstI) | xx µL | xx µL
|
PCR
Tatsui and Yoshida
| KaiC | 10x buffer KOD plus ver2 | 2M dNTP | MgSO4 | Primer Mix(F&R) | Kod-plus ver2 | MilliQ | total
|
| 0.5 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 16.0 | 25.0
|
| 1.0 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 15.5 | 25.0
|
| 2.0 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 14.5 | 25.0
|
| 4.0 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 12.5 | 25.0
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 2min | 30cycles
|
| Plac(100pg) | 10x buffer KOD plus ver2 | 2M dNTP | MgSO4 | Primer Mix(F&R) | Kod-plus ver2 | MilliQ | total
|
| 0.3 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 16.2 | 25.0
|
| 0.6 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 15.9 | 25.0
|
| 1.2 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 15.3 | 25.0
|
| 2.4 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 14.1 | 25.0
|
| 0.0 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 16.5 | 25.0
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 2min | 30cycles
|
Liquid Culture
No name
| Sample | medium
|
| pSB2C3(master)-2 | Plusgrow medium(+CP)
|
Restriction Enzyme Digestion
No name
| | 8/17 lysis1-2 | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 8.1µL | 1µL | 1µL | 3µL | 3µL | 13.9µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 8/17 lysis3-1 | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 2 cuts | 6.1µL | 1µL | 1µL | 3µL | 3µL | 15.9µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 9/15 Plac | XbaI | PstI | BSA | Buffer | MilliQ | total
|
| 1 cuts | 12.1µL | 0µL | 1µL | 3µL | 3µL | 10.9µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
PCR
Yoshida
| KaiA(KaiABC) 100pg/µL | 10x buffer KOD plus ver2 | dNTPs | MgSO4 | Primer Mix(Fwd&Rev) | KOD plus ver2 | MilliQ | total
|
| 2.0 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 14.5 | 25
|
| 4.0 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 12.5 | 25
|
| 8.0 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 8.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 60°C | 68°C | --
|
| 2min | 10s | 30s | 1min | 30cycles
|
| PKaiBC 400pg/µL | 10x buffer KOD plus ver2 | dNTPs | MgSO4 | Primer Mix(Fwd&Rev) | KOD plus ver2 | MilliQ | total
|
| 0.5 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 16.0 | 25
|
| 1.0 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 15.5 | 25
|
| 2.0 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 14.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 60°C | 68°C | --
|
| 2m | 10s | 30s | 1m | 30cycles
|
Liquid Culture
No name
| Sample | medium
|
| 9/2 Spinach-DT | Plusgrow medium(+CP)
|
| 8/29 Plux-RBS-GFP-DT | Plusgrow medium
|
| 9/6 master plate | Plusgrow medium(+Amp)
|
| 3 Pcon-PT181 attenuator | Plusgrow medium
|
No name
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 3 | lysis1 | XbaI+PstI
|
| 4
|
| 5
|
| 6
|
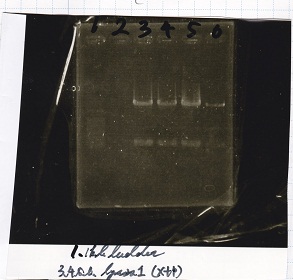

| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 3 | lysis3 | XbaI+PstI
|
| 4
|
| 5
|
| 6
|


| Name | concentration[µg/mL] | 260/280 | 260/230
|
| (´ω`) | | |
|
| (ΦωΦ^) | | |
|
Electrophoresis
Yoshida
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kb ladder | -- | --
|
| 2 | Plac 100pg | -- | --
|
| 3 | Plac 200pg | -- | --
|
| 4 | Plac 400pg | -- | --
|
| 5 | Plac 800pg | -- | --
|
| 6 | NC | -- | --
|
| 7 | 1kbp ladder | -- | --
|

Sep 10
Ligation
Nakamoto
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/6 Pcon-RBS-luxR-DT | 1.9µL | 9/6Plux-RBS-GFP-DT | 22 µL | 3.5µL
|
| experiment | 9/6 Plux | 2µL | 9/8 RBS-lysis1-DT | 6.6µL | 3.5µL
|
| experiment | 9/6 Plux | 2µL | 9/8 RBS-lysis2-DT | 6.3µL | 3.5µL
|
| experiment | 9/8 Ptet-PT181 antisense | 1.6µL | 9/8 spinach-DT | 4.4µL | 3.0µL
|
| experiment | 9/8 Pcon-PT181 attenuator | 3.4µL | 9/8 apz 12-1R-PT | 2.1µL | 2.8µL
|
| experiment | 9/8 Pcon | 3.4µL | 9/8 apz 12-1R-PT | 2.3µL | 2,9µL
|
| experiment | 9/8 DT | 3.0µL | 9/8 Pcon-PT181 attenuator | 7.9µL | 3.5µL
|
| experiment | 9/10 pSB1C3 | 1.8µL | 9/8 F3m2 attenuator | 4.6µL | 3.2µL
|
| experiment | 9/10 pSB1C3 | 1.8µL | 9/8 F1 attenuator | 3.4µL | 2.6µL
|
Miniprep
Nakamoto
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| spinach-DT | 202.4 | 1.97 | 1.93
|
| Plux-RBS-GFP-DT | 164.2 | 2.80 | 2.06
|
| Pcon-DT attenuator | 205.4 | 1.81 | 2.39
|
| RSBJC3-2 | 240.8 | 1.73 | 1.43
|
| Plaz-1 | 240.8 | 1.73 | 1.43
|
Restriction Enzyme Digestion
Nakamoto
| | 9/10 Plux-RBS-GFP-DT | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 2cuts | 12.1µL | 1µL | 1µL | 3µL | 3µL | 9.9µL | 30µL
|
| NC | 0.6µL | 0µL | 0µL | 1µL | 1µL | 7.4µL | 10µL
|
| | 9/10 Spinach-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 9.9µL | 1µL | 1µL | 3µL | 3µL | 12.1µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| | 9/6 apt12-1R -DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 8.0µL | 1µL | 1µL | 3µL | 3µL | 14.0µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 9/10 Pcon-pt81 attenuator | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 2cuts | 9.8µL | 1µL | 1µL | 3µL | 3µL | 12.2µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Plux-RBS-GEP-DT | EcoRI | SpeI
|
| 3 | Plux-RBS-GEP-DT | -- | --
|
| 4 | Spinach-DT | XbaI | PstI
|
| 5 | Spinach-DT | -- | --
|
| 6 | aptamer 121R-DT | XbaI | PstI
|
| 7 | aptamer 121R-DT | -- | --
|
| 8 | Pcon-pt181 attenuator | EcoRI | SpeI
|
| 9 | Pcon-pt181 attenuator | -- | --
|
| 10 | 100bp ladder | -- | --
|

Colony PCR
Tatsui
| Sample | base pair
|
| 9/9 Pcon-pT181 antisense | --
|
| 9/8 J223100(entA) | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min10sec | 30
|
Electrophoresis
Tatsui
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Pcon Pt181 antisense1+spinach-DT
|
| 3 | Pcon Pt181 antisense2+spinach-DT
|
| 4 | Pcon Pt181 antisense3+spinach-DT
|
| 5 | Pcon Pt181 antisense4+spinach-DT
|
| 6 | Pcon Pt181 antisense5+spinach-DT
|
| 7 | Pcon Pt181 antisense6+spinach-DT
|
| 8 | Pcon Pt181 antisense7+spinach-DT
|
| 9 | Pcon Pt181 antisense8+spinach-DT
|
| 10 | Pcon Pt181 antisense9+spinach-DT
|
| 11 | Pcon Pt181 antisense10+spinach-DT
|
| 12 | Pcon Pt181 antisense11+spinach-DT
|
| 13 | Pcon Pt181 antisense12+spinach-DT
|
| 14 | 100bp ladder
|
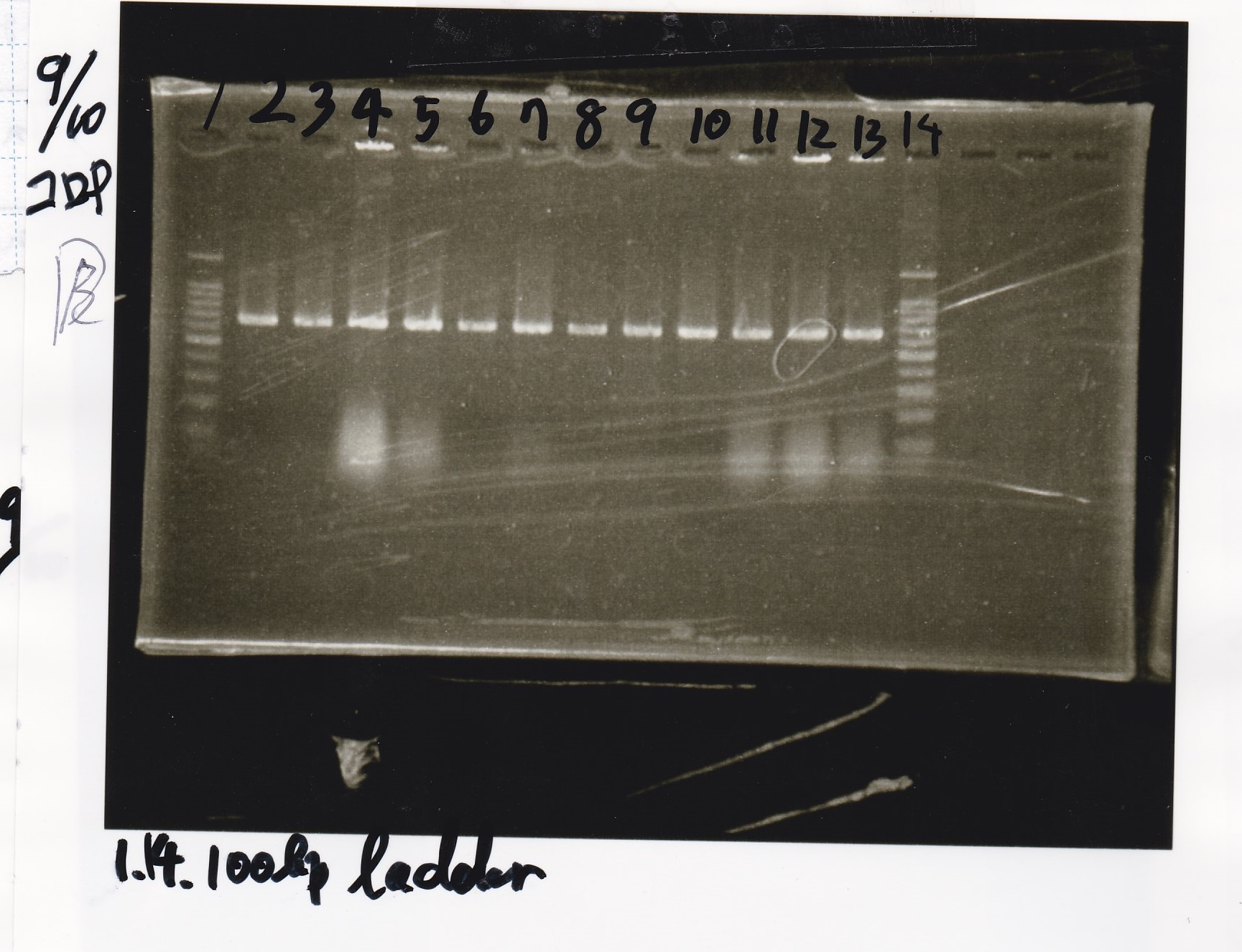
| Lane | Sample
|
| 1 | 1kbp ladder
|
| 2 | Pcon entA1
|
| 3 | Pcon entA2
|
| 4 | Pcon entA3
|
| 5 | 1kbp ladder
|

Tatsui
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 2 | KaiA(200pg) | --
|
| 3 | KaiA(200pg) | --
|
| 4 | -- | --
|
| 5 | KaiA(400pg) | --
|
| 6 | KaiA(400pg) | --
|
| 7 | -- | --
|
| 8 | KaiA(800pg | --
|
| 9 | KaiA(800pg) | --
|
| 10 | 1kbp ladder | --
|


| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 2 | PKaiBC(200pg) | --
|
| 3 | PKaiBC(200pg) | --
|
| 4 | -- | --
|
| 5 | PKaiBC(400pg) | --
|
| 6 | PKaiBC(400pg) | --
|
| 7 | -- | --
|
| 8 | PKaiBC(800pg | --
|
| 9 | PKaiBC(800pg) | --
|
| 10 | 1kbp ladder | --
|
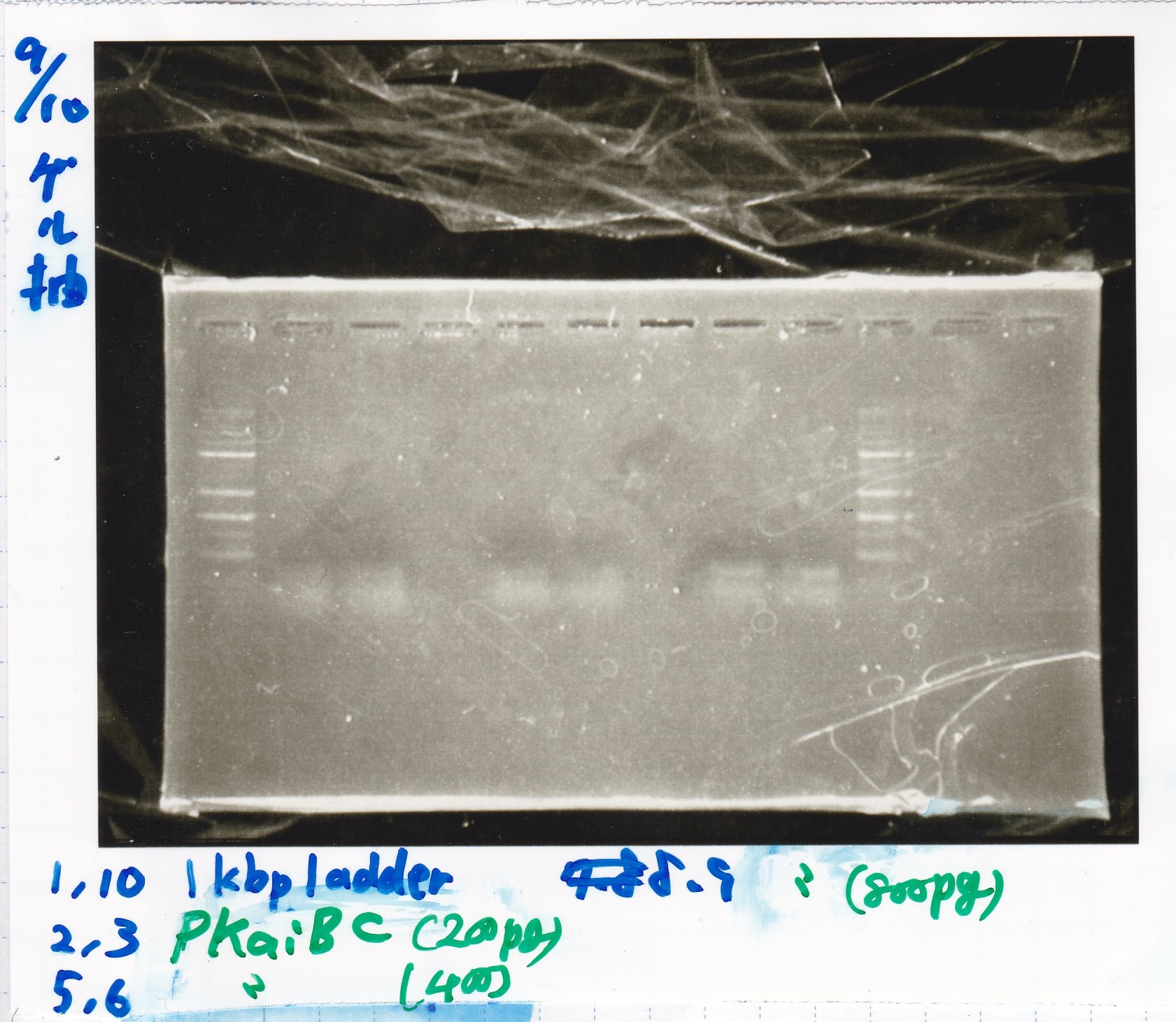
Liquid Culture
Hirano
| Sample | medium
|
| 9/4 J23100①(JM109) | LB(Amp)
|
| 9/8 J23100① (entA) | LB(Amp)
|
Tatsui
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 2 | KaiC | --
|
| 3 | KaiC | --
|
| 4 | KaiC | --
|
| 5 | -- | --
|
| 6 | 1kbp ladder | --
|
| 7 | 1kbp ladder | --
|
| 8 | KaiC | --
|
| 9 | KaiC | --
|
| 10 | KaiC | --
|
| 11 | -- | --
|
| 12 | 1kbp ladder | --
|
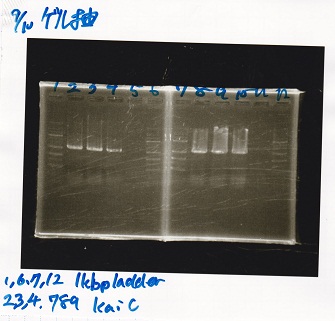

Liquid Culture
Tatsui
| Sample | medium
|
| 9/10 Pcon-pt181 antisense-Spinach-DT | Plusgrow
|
Colony PCR
No name
| Sample | base pair
|
| 9/9 Phad larac-1 RBS-lux1-DT | 2331
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 2min | 30
|
| Sample | base pair
|
| 9/9 Pcon-lacZα (psB4KS) | 712
|
| 9/9 Pcon-Spinach-DT(psB4KS) | 605
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 42s | 30
|
| Sample | base pair
|
| 9/9 Plac+RBS-lysis3}-DT | 1273
|
| 9/9 Pcon+RBS-lux1-DT | 1079
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min12s | 30
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | 9/9 Pcou-RBS-lacZα-DT(p3B4KS9)-1 | -- | --
|
| 3 | 9/9 Pcou-RBS-lacZα-DT(p3B4KS9)-2 | -- | --
|
| 4 | 9/9 Pcou-Spinach-DT(pB4KS)-1 | -- | --
|
| 5 | 9/9 Pcou-Spinach-DT(pB4KS)-2 | -- | --
|
| 6 | 100bp ladder | -- | --
|

Transformation
Nakamoto
| Name | Plate
|
| F1 attenuator +p3B1C3 | CP
|
| F3n2 attenuator +p3B1C3 | CP
|
| F1 antisense +p3B1C3 | CP
|
| F6 antisense+p3B1C3 | CP
|
| apt12-P +p3B1C3 | CP
|
| apt12-1M +p3B1C3 | CP
|
| Pcon-luxR+Plux-GFP | Amp
|
| Plux+RBS-lysis1-DT | CP
|
| Plux+RBS-lysis2-DT | CP
|
| Ptet-pT181antisense+Spinach-DT | CP
|
| Pcon-pT181 attenuator+apt121R-DT | Amp
|
| Pcon-1 apt12-1R-DT | Amp
|
| DT+Pcon-pT181 attenuator | CP
|
Electrophoresis
Okazaki
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Spinach-DT | XbaI | PstI
|
| 3 | Spinach-DT | XbaI | PstI
|
| 4 | Spinach-DT | XbaI | PstI
|
| 5 | Spinach-DT | XbaI | PstI
|
| 6 | 100bp ladder | -- | --
|
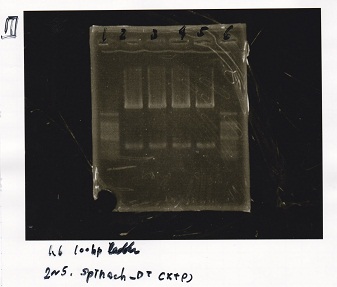

| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Pcon-pT181 attenuator | EcoRI | SpeI
|
| 3 | Pcon-pT181 attenuator | EcoRI | SpeI
|
| 4 | Pcon-pT181 attenuator | EcoRI | Spei
|
| 5 | Pcon-pT181 attenuator | EcoRI | SpeI
|
| 6 | 100bp ladder | -- | --
|
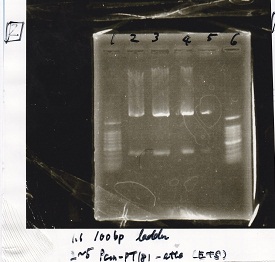
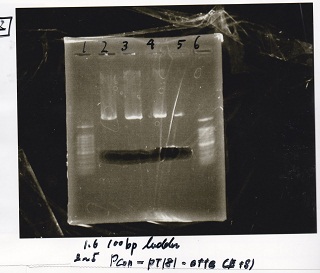
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | Plux-RBS-GFP-DT | EcoRI | SpeI
|
| 2 | Plux-RBS-GFP-DT | EcoRI | SpeI
|
| 3 | Plux-RBS-GFP-DT | EcoRI | SpeI
|
| 4 | Plux-RBS-GFP-DT | EcoRI | SpeI
|
| 5 | 100bp ladder | -- | --
|
| 6 | apt12-1R-DT | XbaI | PstI
|
| 7 | apt12-1R-DT | XbaI | PstI
|
| 8 | apt12-1R-DT | XbaI | PstI
|
| 9 | apt12-1R-DT | XbaI | PstI
|
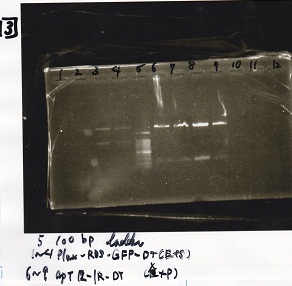
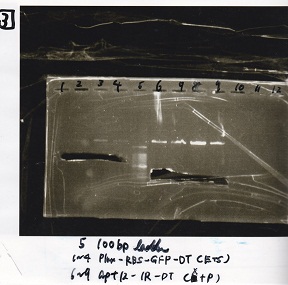
Sep 11
Electrophoresis
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | Pbad/araC+RBS-luxI-DT-1
|
| 3 | Pbad/araC+RBS-luxI-DT-1
|
| 4 | Plac+RBS-lysis-DT
|
| 5 | Pcon+RBS-luxI-DT
|
| 6 | 1kb ladder
|

PCR
Tatsui
| pKaiBC(200p) | 10*buffer KOD plus | dNTPs | MgSO4 | primer mix(F&R) | KOD plus | MilliQ | total
|
| 0.5 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 16 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 60°C | 68°C | 30
|
| 2min | 10sec | 30sec | 30sec |
|
| pKaiBC(400p) | 10*buffer KOD plus | dNTPs | MgSO4 | primer mix(F&R) | KOD plus | MilliQ | total
|
| 1 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 60°C | 68°C | 30
|
| 2min | 10sec | 30sec | 30sec |
|
| pKaiBC(800p) | 10*buffer KOD plus | dNTPs | MgSO4 | primer mix(F&R) | KOD plus | MilliQ | total
|
| 2 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 14.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 60°C | 68°C | 30
|
| 2min | 10sec | 30sec | 30sec |
|
| pKaiBC(1ng) | 10*buffer KOD plus | dNTPs | MgSO4 | primer mix(F&R) | KOD plus | MilliQ | total
|
| 2.5 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 14 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 60°C | 68°C | 30
|
| 2min | 10sec | 30sec | 30sec |
|
Electrophoresis
Tatsui
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | pKaiBC 800pg 60°C
|
| 3 | pKaiBC 1ng 60°C
|
| 4 | pKaiBC 800pg 61°C
|
| 5 | pKaiBC 1ng 61°C
|
| 6 | pKaiBC 800pg 62°C
|
| 7 | pKaiBC 1ng 62°C
|
| 8 | 100bp ladder
|
colony PCR
Tatsui
| Sample | base pair
|
| 9/9Pcon-lacZα(p3B4k5) | 712bp
|
| 9/9Pcon-spinach-DT(p3B4k5) | 605bp
|
Tatsui
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 42s | 30
|
| Sample | base pair
|
| Pbad\araC-RBS-luxI-DT |
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 2min | 30
|
Electrophoresis
Tatsui
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Pcon-lacZα(pSB4K5)3
|
| 3 | Pcon-lacZα(pSB4K5)4
|
| 4 | Pcon-lacZα(pSB4K5)5
|
| 5 | Pcon-lacZα(pSB4K5)6
|
| 6 | Pcon-spinach-DT(pSB4K5)3
|
| 7 | Pcon-spinach-DT(pSB4K5)4
|
| 8 | Pcon-spinach-DT(pSB4K5)5
|
| 9 | Pcon-spinach-DT(pSB4K5)6
|
| 10 | 100bp ladder
|
| Lane | Sample
|
| 1 | 1kbp ladder
|
| 2 | Pbad/araC-RBS-luxI-DT3
|
| 3 | Pbad/araC-RBS-luxI-DT4
|
| 4 | Pbad/araC-RBS-luxI-DT5
|
| 5 | Pbad/araC-RBS-luxI-DT6
|
| 6 | 1kbp ladder
|

colony PCR
| Sample | base pair
|
| 9/9Pcon+RBS-luxI-DT | 1079bp
|
| 9/10Pcon+sptamer12_1R-DT | 584bp
|
| 9/10DT+Pcon-pT181 attenuator 1 | 781bp
|
| 9/10Fusion1 attenuator(p3B1C3)-1 | 638bp
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 50sec | 30sec | 30sec | 1min | 30
|
Ligation
Okazaki
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/8pSB4K5(EcoRI&SpeI)21.8ng/µL | 2.3 µL | 9/8Pcon-Spinach-DT(EcoRI&SpeI)10.5ng/µL | 1.7 µL | 2.0 µL
|
| experment | 9/8pSB4K5(EcoRI&SpeI)21.8ng/µL | 2.3 µL | 9/8RBS-lacZα-DT(EcoRI&SpeI)4.4ng/µL | 6.7µL | 3.5 µL
|
| experiment | 9/8pSB4K5(EcoRI&SpeI)21.8ng/µL | 2.3 µL | 9/8Pcon-RBS-lacZα-DT(EcoRI&SpeI)5.6ng/µL | 1.9 µL | 2.1 µL
|
| experiment | 9/6Plux 25ng/µL | 2 µL | 9/8 RBS-lysis1-DT(XbaI&PstI) | 6.6 µL | 3.5 µL
|
| experiment | 9/6Plux 25ng/µL | 2 µL | 9/8 RBS-lysis3-DT(XbaI&PstI) | 4.7 µL | 3.4 µL
|
| experiment | 9/10Pcon-pT181 attenuator(SpeI&PstI) | 3.4µL | 9/8RBS-lacZα-DT(EcoRI&SpeI) | 4.5 µL | 3.5 µL
|
| experiment | 9/6Pcon-RBS laxR-DT(EcoRI&XbaI) 26.7ng/µL | 1.9 µL | Plax-RBS-GFP-DT | 5.5 µL | 3.5 µL
|
| experiment | 9/9 Pcon-pT181 attenuator(SpeI&PstI) | 3.4 µL | 9/10 aptamer12_1R-DT(XbaI&PstI) | 1.7 µL | 2.6 µL
|
Transformation
Nakamoto Tatsui
| Name | Sample | Competent Cells | Total | Plate
|
| 9/9 Fusion1 antisense(pSB2C3) | 2µL | 20µL | 22µL | CP
|
| 9/9 Fusion6 antisense(pSB2C3) | 2µL | 20µL | 22µL | CP
|
| 9/9 aptamer 12_P(pSB2C3) | 2µL | 20µL | 22µL | CP
|
| 9/9 aptamer 12_1µ(pSB2C3) | 2µL | 20µL | 22µL | CP
|
| 9/9 Fusion2 attenuator(pSB2C3) | 2µL | 20µL | 22µL | CP
|
| 9/11 Pcon-spinach-DT(pSB4K5) | 2µL | 20µL | 22µL | K
|
| 9/11 RBS-lacZα-DT(pSB4K5) | 2µL | 20µL | 22µL | K
|
| 9/11 Pcon-RBS-lacZα-DT(pSB4K5) | 2µL | 20µL | 22µL | K
|
| Plux+RBS-lysis1-DT | 2µL | 20µL | 22µL | CP
|
| Plux+RBS-lysis3-DT | 2µL | 20µL | 22µL | CP
|
| Pcon-pT181attenuator+RBS-lacZα-DT | 2µL | 20µL | 22µL | Amp
|
| Pcon-iuxR+Plux-GFP | 2µL | 20µL | 22µL | Amp
|
| Pcon-pT181attenuator+apt12_1R-DT | 2µL | 20µL | 22µL | Amp
|
| Plac(BBa_R0011) | 2µL | 20µL | 22µL | Amp |
|
Electrophoresis
Tatsui
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | Pcon+RBS luxI-DT2
|
| 3 | Pcon+RBS luxI-DT3
|
| 4 | Pcon+RBS luxI-DT4
|
| 5 | Pcon+RBS luxI-DT5
|
| 6 | Pcon+RBS luxI-DT6
|
| 7 | Pcon+apF12_1R-DT1
|
| 8 | Pcon+apF12_1R-DT2
|
| 9 | Pcon+apF12_1R-DT3
|
| 10 | Pcon+apF12_1R-DT4
|
| 11 | DT+Pcon-pT181attenuator1
|
| 12 | F1attenuator(pSB1C3)
|
| 13 | 100bp ladder
|
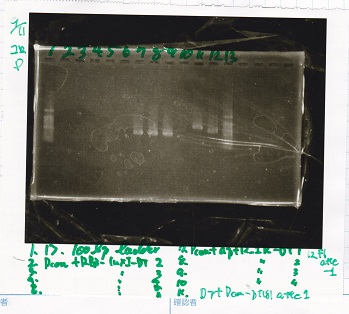
Restriction Enzyme Digestion
Nakamoto
| | DNA | EcoRI | XbaI | SpeI | PstI | buffer | BSA | MilliQ | Total
|
| pSB4K5(EcoRI+SpeI) | 9.4µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 12.6µL | 30 µL
|
| pSB4K5(EcoRI+SpeI) negative | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| RBS-lacZα-DT(EcoRI+SpeI) | 10µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 12µL | 30µL
|
| RBS-lacZα-DT(EcoRI+SpeI) negative | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| Ptet-pT181 antisense(SteI+PstI) | 10.8µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 12µL | 30µL
|
| Ptet-pT181 antisense(SteI+PstI) negative | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| Pcon-RBS-lacZα-DT(EcoRI+SpeI) | 7.1µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 14.9µL | 30µL
|
| Pcon-RBS-lacZα-DT(EcoRI+SpeI) negative | 0.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| RBS-lysis2-DT(XbaI+PstI) | 22µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 0µL | 30µL
|
| RBS-lysis2-DT(XbaI+PstI) negative | 1.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 6.6µL | 10µL
|
Liquid Culture
Nakamoto
| Sample | medium
|
| 8/16 Master4 Plax |
|
| 8/25 Pλ-laxI-2 |
|
PCR
Hirano
| 8/29 Pλ-RBS-luxI-DT(76.5ng/µL) | 10x buffer for KOD plus-ver.2 | KOD-plus | dNTP | MgSO4 | F primer(RBS-luxI-DT-cloning-fwd) | R primer(RBS-luxI-DT-cloning-rev) | MilliQ | total
|
| 0.3 | 2.5 | 0.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.2 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 65°C | 68°C | --
|
| 5min | 30sec | 30sec | 1min | 30
|
| 8/31 Pbad/araC-RBS-RFP(1)(67.6ng/µL) | 10x buffer for KOD plus-ver.2 | KOD-plus | dNTP | MgSO4 | F primer(Pbad/araC(pSBlc3)-cloning-fwd) | R primer(Pbad/araC(pSBlc3)-cloning-rev) | MilliQ | total
|
| 0.3 | 2.5 | 0.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.2 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 54°C | 68°C | --
|
| 5min | 30sec | 30sec | 3min 20sec | 30
|
Liquid Culture
Hirano
| Sample | medium
|
| 9/4 RBS-lysis1+DT 3 | plusgrow(+CP) 3ml 37°C
|
| 9/4 RBS-lysis2+DT 1 | plusgrow(+CP) 3ml 37°C
|
| 9/4 RBS-lysis3+DT 3 | plusgrow(+CP) 3ml 37°C
|
Sep 12
Miniprep
Nakamoto
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-pT181attenuator-DT | 293.6 | 1.88 | 1.84
|
| Pcon-apt12-1R-DT | 303.8 | 1.75 | 1.64
|
| Fusion1attenuator(pSB1c3) | 172.0 | 1.96 | 2.31
|
| RBS-lisis1-DT | 235.1 | 1.59 | 1.60
|
| RBS-lisis2-DT | 387.7 | 1.91 | 2.01
|
| RBS-lisis3-DT | 387.9 | 1.91 | 2.09
|
| Plux | 166.7 | 1.98 | 1.63
|
Electrophoresis
Nakamoto
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | 9/11 P-RBS-lisis1-DT(Colony PCR prodution)
|
Liquid Culture
Nakamoto
| Sample | medium
|
| 9/10 Pcon-apt12-1R-DT1 | Plusgrow medium(+Amp)
|
| 9/10 DT-Pcon-pT181attenuator1 | Plusgrow medium(+CP)
|
| 9/10 Fusion1-attenuator(pSB1c3) | Plusgrow medium(+CP)
|
Electrophoresis
Tatsui
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | pSB4K5(EcoRI&SpeI)
|
| 3 | pSB4K5 NC(EcoRI&SpeI)
|
| 4 | RBS-lacz-DT(EcoRI&SpeI)
|
| 5 | EcoRI&SpeI NC( EcoRI&SpeI)
|
| 6 | Pcon-RBS-lacz-DT( EcoRI&SpeI)
|
| 7 | Pcon-RBS-lacz-DT NC( EcoRI&SpeI)
|
| 8 | 1kb ladder
|
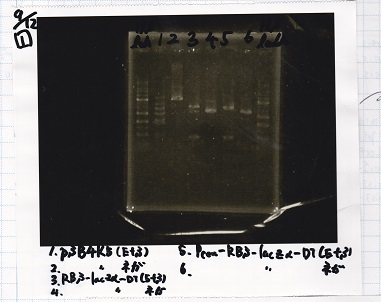
2
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | Ptet-pT181antisense(SpeI&PstI)
|
| 3 | Ptet-pT181antisense NC(SpeI&PstI)
|
| 4 | RBS-lysis2-DT(XbaI&PstI)
|
| 5 | EcoRI&SpeI NC( EcoRI&SpeI)
|
| 6 | --
|
| 7 | --
|
| 8 | 1kb ladder
|
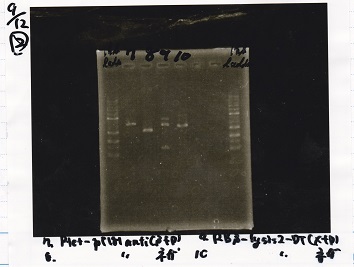
Tatsui
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 2 | RBS-lacZα-DT | EcoRI&SpeI
|
| 3 | RBS-lacZα-DT | EcoRI&SpeI
|
| 4 | RBS-lacZα-DT | EcoRI&SpeI
|
| 5 | -- | --
|
| 6 | Pcon-RBS-lacZα-DT | EcoRI&SpeI
|
| 7 | Pcon-RBS-lacZα-DT | EcoRI&SpeI
|
| 8 | Pcon-RBS-lacZα-DT | EcoRI&SpeI
|
| 9 | 1kb ladder | --
|
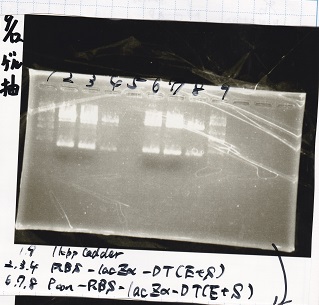
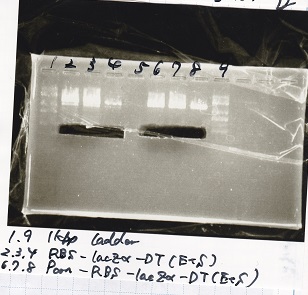
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-lacZα-DT(EcoRI&SpeI) | 9.8 | 1.50 | 0.39
|
| Pcon-RBS-lacZα-DT(EcoRI&SpeI) | 11.2 | 1.47 | 0.08
|
Tatsui
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 2 | RBS-lysis2-DT | XbaI&PstI
|
| 3 | RBS-lysis2-DT | XbaI&PstI
|
| 4 | RBS-lysis2-DT | XbaI&PstI
|
| 5 | -- | --
|
| 6 | 1kb ladder | --
|

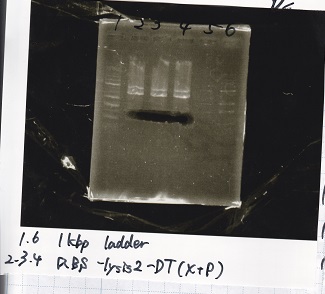
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-lysis2-DT(XbaI&PstI) | 5.4 | 1.43 | 0.77
|
Electrophoresis
No name
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | Pλ-RBS-luxI-DT(PCR product)
|
| 3 | Pbad/araC-RBS-RFP(PCR product)
|

Colony PCR
Honda
| Sample | base pair
|
| 9/11 apt12-1M(pSB1C3) 1 | 513
|
| 9/11 apt12-1M(pSB1C3) 2 | 513
|
| 9/11 Fusion3m2a09ttenuator(pSB1C3) 1 | 609
|
| 9/11 Fusion6 antisense(pSB1C3)1 | 431
|
| 9/11 aptamer 12-P(pSB1C3)1 | 515
|
| 9/11 Fusion1 antisense(pSB1C3) 1 | 420
|
| 9/11 Fusion1 antisense(pSB1C3) 2 | 420
|
| 9/11 Plac(BBa-R0011) 1 | 293
|
| 9/11 Plac(BBa-R0011) 2 | 293
|
| 9/11 Plac(BBa-R0011) 3 | 293
|
| 9/11 Plac(BBa-R0011) 4 | 293
|
| 9/11 Pcon-luxR-Plux-GFP 1 | 2138
|
| 9/11 Pcon-RBS-lacZα-DT(pSB4K5) 1 | 712
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 2min12s | 30cycles
|
Restriction Enzyme Digestion
Tatsui
| | 9/5 Pcon | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cuts(PstI) | 5.7 | 0µL | 0µL | 0µL | 1µL | 3µL | 3µL | 17.3µL | 30µL
|
| NC(PstI) | 1.9 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 6.1µL | 10µL
|
| | 8/17 DT | EcoRI | Buffer | BSA | MilliQ | total
|
| 1cut(EcoRI) | 10.6 | 1µL | 3µL | 3µL | 12.4µL | 30µL
|
| NC(EcoRI) | 3.5 | 0µL | 1µL | 1µL | 4.5µL | 10µL
|
Colony PCR
Nakamoto and Tatsui
| Sample | base pair
|
| 9/11 Pcon-pT181attenuator-RBS-lacZα-DT | 965
|
| 9/11 Plux-PBS-lysis1-DT 1 | 613
|
| 9/11 Pcon-pT181attenuator-aptamer12-1R-DT 1 | 859
|
| 9/11 Pcon-pT181attenuator-aptamer12-1R-DT 2 | 859
|
| 9/11 Plux-RBS-lysis3-DT 1 | 1210
|
| 9/11 Plux-RBS-lysis3-DT 2 | 1210
|
| Pcon-Spinach-DT(pSB4K5) 1 | 605
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min12s | 30cycles
|
Transformation
Tatsui
| Name | Sample | Competent Cells | Plate
|
| 9/7 pSB4K5 | 2µL | 20µL | Amp
|
Electrophoresis
No name
1
| Lane | Sample
|
| 1 | Fusion6 antisense
|
| 2 | Fusion1 antisense-1
|
| 3 | Fusion1 antisense-2
|
| 4 | 100bp ladder
|
| 5 | Plac(BBa-R0011) 1
|
| 6 | Plac(BBa-R0011) 2
|
| 7 | Plac(BBa-R0011) 3
|
| 8 | Plac(BBa-R0011) 4
|

2
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | aptamer12-1M 1
|
| 3 | aptamer12-1M 2
|
| 4 | Fusion3m2 attenuator
|
| 5 | aptamer12-P1
|
| 6 | Plux-RBS-GFP-DT-Pcon-RBS-luxR-DT
|
| 7 | Pcon-RBS-lacZα=DT
|
| 8 | 1kb ladder
|
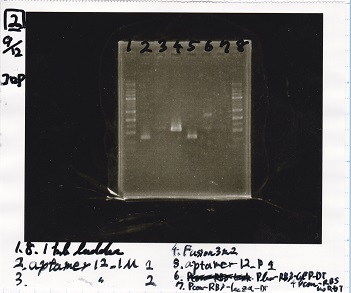
Electrophoresis
No name
| Lane | Sample | Enzyme
|
| 1 | Pcon-pT181attenuator-RBS-lacZα-DT 1 | --
|
| 2 | Pcon-pT181attenuator-RBS-lacZα-DT 2 | --
|
| 3 | Plux-RBS-lysis1-DT 1 | --
|
| 4 | Pcon-pT181attenuator-aptamer12-1R-DT 1 | --
|
| 5 | Pcon-pT181attenuator-aptamer12-1R-DT 2 | --
|
| 6 | Plux-RBS-lysis3-DT 1 | --
|
| 7 | 1kbp ladder | --
|
| 8 | Plux-RBS-lysis3-DT 2 | --
|
| 9 | Pcon-spinach-DT(pSB4K5) 1 | --
|
| 10 | 9/5 Pcon | PstI
|
| 11 | 9/5 Pcon NC | --
|
| 12 | 8/17 DT | EcoRI
|
| 13 | 8/17 DT | --
|

Liquid Culture
Nakamoto
| Sample | medium
|
| 9/11Fusion6 antisense-1 | Plusgrow medium(+CP)
|
| Plac(BBa-R0011)-3 | Plusgrow medium(+Amp)
|
| Plac(BBa-R0011)-4 | Plusgrow medium(+Amp)
|
| Fusion3m2 attenuator -1 | Plusgrow medium(+CP)
|
| Pλ-luxI-2 | Plusgrow medium(+Amp)
|
| 9/11 aptamer12-1M-2 | Plusgrow medium(+CP)
|
Colony PCR
Nakamoto
| Sample | base pair
|
| 9/11 Fusion1 antisense 3 | 394
|
| 9/11 Fusion1 antisense 4 | 394
|
| 9/11 aptamer12-P 2 | 378
|
| 9/11 aptamer12-P 3 | 378
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 36s | 30cycles
|
Electrophoresis
Nakamoto
| Lane | Sample | Enzyme
|
| 1 | Fusion1 antisense 2 | --
|
| 2 | Fusion1 antisense 3 | --
|
| 3 | 1kb ladder | --
|
| 4 | apt12-P 2 | --
|
| 5 | apt12-P 3 | --
|
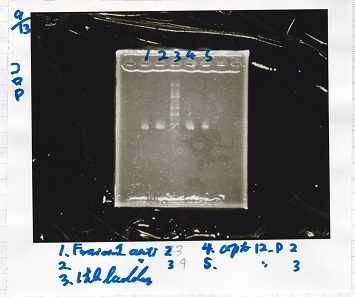
Restriction Enzyme Digestion
Tatsui
| | 9/12 RBS-lysis2-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 5.2 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 16.8µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.7µ | 10µL
|
| | 9/12 RBS-lysis3-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 5.2 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 16.8µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.7µ | 10µL
|
| | 9/11 RBS-lysis1-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 8.5 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 13.5µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.6µ | 10µL
|
| | 8/20 J23100-RBS-luxR -DT | EcoRI | Buffer | BSA | MilliQ | total
|
| 1cut | 5.8 | 1.0µL | 3.0µL | 3.0µL | 17.2µL | 30µL
|
| NC | 0.3µL | 0µL | 1.0µL | 1.0µL | 7.7µ | 10µL
|
| | 9/10 pSB1C3 | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 2cuts | 8.1 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 13.9µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.6µ | 10µL
|
| | 9/12 Plux | PstI | Buffer | BSA | MilliQ | total
|
| 1cut | 12.0 | 1.0µL | 3.0µL | 3.0µL | 11.0micro;L | 30µL
|
| NC | 0.6µL | 0µL | 1.0µL | 1.0µL | 7.4µ | 10µL
|
| | 9/10 pSB1C3 | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 8.1 | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 13.9µL | 30µL
|
Electrophoresis
Tatsui
| Lane | Sample | Enzyme1 | Enzymel2
|
| 1 | 9/12 RBS-lysis2-DT | XbaI | PstI
|
| 2 | 9/12 RBS-lysis2-DT NC | -- | --
|
| 3 | 9/11 RBS-lysis1-DT | XbaI | PstI
|
| 4 | 9/11 RBS-lysis1-DT NC | -- | --
|
| 5 | 9/11 RBS-lysis3-DT | XbaI | PstI
|
| 6 | 9/11 RBS-lysis3-DT NC1 | -- | --
|
| 7 | 1kb ladder | -- | --
|
| 8 | 100bp ladder | -- | --
|
| 9 | J23100-RBS-luxR-DT | EcoRI | --
|
| 10 | J23100-RBS-luxR-DT | -- | --
|
| 11 | 9/10 pSB1C3 | EcoRI | SpeI
|
| 12 | 9/10 pSB1C3 NC | -- | ---
|
| 13 | 9/10 pSB1C3 | XbaI | PstI
|
| 14 | 9/12 Plux | PstI | --
|
| 15 | 9/12 Plux NC | -- | --
|
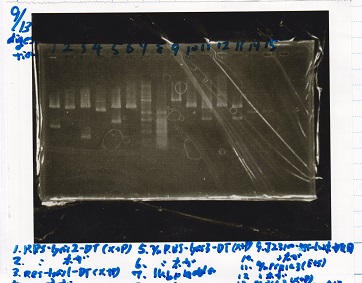
Observation through a confocal microscope
We used the liquid culture media with 200μL 9/10Pcon-pT181antisense-spinach-DT& 200μL 9/10 Pcon-spinach-DT& 200μL 9/10 JM109(overnight culture).
We translated them into each 1.5mL tube.
We elminated each supernatant using a 5000xg centrifuge for 1minute,.
Then,we resuspended with 100µL M9(distilled water) and eliminated each supernatant using a 5000xg centrifuge for 1 minute.x2
Adding 100µL M9(in 20µM DFHBI), we resuspended the pret.
15min after incubating in shield light, we eliminated each supernatant using a 5000xg centrifuge for 1 minute.
We looked at the fluorescence of the pret.
We failed to observe it.
Liquid Culture
Hirano
| Sample | medium
|
| 9/10 Pcon-pT181antisense-spinach-DT | Plusgrow medium
|
| 9/10 Pcon-spinach-DT | Plusgrow medium
|
| 9/1 spinach-DT(RNA master plate) | Plusgrow medium
|
Sep 13
Miniprep
Nakamoto
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/12 Fusion6 antisense | 245.9 | 1.88 | 1.41
|
| 9/12 Plux-3 | 115.5 | 1.58 | 1.32
|
| 9/12 Plux-4 | 125.3 | 1.93 | 1.44
|
| 9/12 Fusion3n2 attenuator-1 | 231.9 | 1.81 | 1.31
|
Tatsui,Ashida
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 2 | RBS-lysis1-DT | XbaI&PstI
|
| 3 | RBS-lysis1-DT | XbaI&PstI
|
| 4 | RBS-lysis1-DT | XbaI&PstI
|

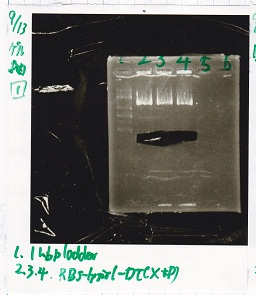
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 3 | RBS-lysis2-DT | XbaI&PstI
|
| 4 | RBS-lysis2-DT | XbaI&PstI
|
| 5 | RBS-lysis2-DT | XbaI&PstI
|
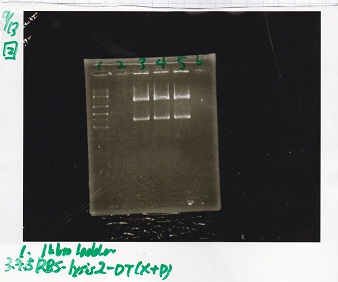

| Lane | DNA | Enzyme
|
| 3 | RBS-lysis3-DT | XbaI&PstI
|
| 4 | RBS-lysis3-DT | XbaI&PstI
|
| 5 | RBS-lysis3-DT | XbaI&PstI
|
| 6 | 1kb ladder | --
|
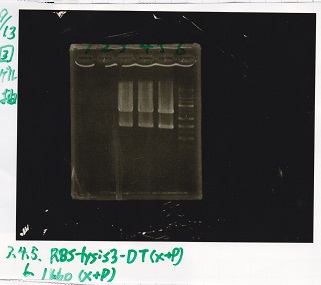
| Lane | DNA | Enzyme
|
| 2 | 1kb ladder | --
|
| 3 | pSB1C3 | EcoRI&SpeI
|
| 4 | pSB1C3 | EcoRI&SpeI
|
| 5 | pSB1C3 | EcoRI&SpeI
|


| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 2 | pSB1C3 | XbaI&PstI
|
| 3 | pSB1C3 | XbaI&PstI
|
| 4 | pSB1C3 | XbaI&PstI
|
Restriction Enzyme Digestion
Nakamoto
| | 9/13 Plac | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cuts(PstI) | 16 | 0µL | 0µL | 0µL | 1µL | 3µL | 3µL | 7µL | 30µL
|
| NC | 0.8 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.2µL | 10µL
|
| | 9/10 Pcon attenuator | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cut(PstI) | 10 | 0µL | 0µL | 0µL | 1µL | 3µL | 3µL | 13µL | 30µL
|
| NC | 0.5 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| | 9/12 Plux | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cuts(PstI) | 12 | 0µL | 0µL | 0µL | 1µL | 3µL | 3µL | 11µL | 30µL
|
| NC | 0.6 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.4µL | 10µL
|
| | 9/12 DT-E | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cut(XbaI) | 23.7 | 0µL | 0µL | 1µL | 0µL | 3µL | 3µL | 0µL | 30.7µL
|
| NC | 1.2 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 6.8µL | 10µL
|
| | 9/12 Pcon-RBS-luxR-bTE | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cuts(XbaI) | 22.4 | 0µL | 0µL | 1µL | 0µL | 3µL | 3µL | 0.6µL | 30µL
|
| NC | 2.2 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 5.8µL | 10µL
|
| | 9/12 Pcon-P | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cut(SpeI) | 21 | 0µL | 1µL | 0µL | 0µL | 3µL | 3µL | 12.0µL | 30µL
|
| NC | 2.1 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 5.7µL | 10µL
|
M9 (EDTA)
| volume | 10ml
|
| Na2HPO4 | 300mg
|
| KH2PO4 | 150mg
|
| NaCl | 25mg
|
| NH4Cl | 50mg
|
| Fe(3)-EDTA | 21mg
|
5mM EDTA
| volume | 10ml
|
| Na2HPO4 | 300mg
|
| KH2PO4 | 150mg
|
| NaCl | 25mg
|
| NH4Cl | 50mg
|
| Fe(3)-EDTA | 105mg
|
Nakamoto
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 3 | Pcon-P | SpeI
|
| 4 | Pcon-P | SpeI
|
| 5 | Pcon-P | SpeI
|


Liquid Culture
Tatsui
| Sample | medium
|
| 9/11 aptamer12_1M(pSB1C3)-1 | --
|
| 9/11 aptamer 12_P1 | --
|
| 9/11 Fusaion1 antisense-4 | --
|
| 9/12 pSB4K5-1 | --
|
| 9/12 pSB4K5-2 | --
|
Restriction Enzyme Digestion
Nakamoto
| | 9/13 Plac-P | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cuts(SpeI) | 23.5 | 0µL | 0.5µL | 0µL | 0µL | 3µL | 3µL | 0µL | 30µL
|
| NC | 8 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 0µL | 10µL
|
| | 9/13 Pcon-pT181 attenuator-P | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cut(SpeI) | 23.5 | 0µL | 0.5µL | 0µL | 0µL | 3µL | 3µL | 0µL | 30µL
|
| NC | 02.3 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 5.7µL | 10µL
|
| | 9/13 Plux-P | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 1cuts(SpeI) | 23.8 | 0µL | 0.2µL | 0µL | 1µL | 3µL | 3µL | 0µL | 30µL
|
| NC | 8 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 0µL | 10µL
|
| | 9/5 RBS-lacZα-DT | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cut(XbaI&PstI) | 10.8 | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 11.2µL | 30µL
|
| NC | 0.5 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
Electrophoresis
Hirano
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | RBS-lacZα-DT(XbaI&PstI)
|
| 3 | RBS-lacZα-DT(NC)
|
Ligation
No Name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/13 Plac (SpeI&PstI) | 8.5µL | 9/13 pT181 antisense (XbaI&PstI) | 2.1 µL | 5.3 µL
|
| experiment | 9/13 Plac (SpeI&PstI) | 8.5 µL | 9/10 aptamer 12_1R-DT (XbaI&PstI) | 1.8 µL | 5.15µL
|
| experiment | 9/13 Plac (SpeI&PstI) | 8.5 µL | 9/2 pT181 attenuator(XbaI&PstI) | 2.3 µL | 10.4 µL
|
| experiment | 9/13 Plux (SpeI&PstI) | 11.4 µL | 9/13 RBS-lysis1-DT (XbaI&PstI) | 5.0 µL | 8.2 µL
|
| experiment | 9/13 Plux (SpeI&PstI) | 11.4 µL | 9/13 RBS-lysis2-DT (XbaI&PstI) | 7.0 µL | 9.2 µL
|
| experiment | 9/13 Plux (SpeI&PstI) | 7.2 µL | 9/13 RBS-lysis3-DT (XbaI&PstI) | 6.0 µL | 6.6 µL
|
| experiment | 9/13 Pcon-RBS-luxR-DT (EcoRI&XbaI) | 2.0 µL | 9/4 Plux-RBS-GFP-DT (EcoRI&SpeI) | 5.2 µL | 3.6 µL
|
| experiment | 9/13 Pcon-RBS-luxR-DT (EcoRI&XbaI) | 2.0 µL | 9/10 aptamer 12_1R-DT (XbaI&PstI) | 1.6 µL | 1.8 µL
|
| experiment | 9/12 Ptet-pT181 antisense (--) | 1.0 µL | 9/10 spinach-DT (--) | 2.4 µL | 1.7 µL
|
PCR
No name
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/7 tetR aptamer 12_1R(pSB1C3) | 0.5 | 6.05 | 1.75 | 1.7 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/6 pT181 antisense-1(pSB1C3) | 0.5 | 6.05 | 1.75 | 1.7 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/6 pT181 attenuator (pSB1C3) | 0.5 | 3.85 | 1.75 | 3.9 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 8/27 Pλ-RBS-luxI-DT-1 | 0.5 | 2.55 | 1.75 | 5.2 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 8/31 Pbad/araC-RBS-RFP-2 | 0.5 | 3.75 | 1.75 | 4 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/13 Plac(pSB1A2) | 0.5 | 4.2 | 1.75 | 3.5 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 8/20 J23100-RBS-lacZα-DT | 0.5 | 6.35 | 1.75 | 1.4 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/7 Ptet-RBS-lacZα-DT | 0.5 | 6.05 | 1.75 | 1.7 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/8 J23100-spinach-DT | 0.5 | 5.75 | 1.75 | 2 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/11 RBS-lysis1-DT | 0.5 | 6.05 | 1.75 | 1.7 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/12 RBS-lysis2-DT | 0.5 | 6.75 | 1.75 | 1.0 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/12 RBS-lysis3-DT | 0.5 | 6.75 | 1.75 | 1.0 | 0.5 | 0 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/8 J23100-spinach-DT | 0.5 | 6.35 | 1.75 | 1.4 | 0 | 0.5 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 8/27 Pλ-RBS-luxI-DT-1 | 0.5 | 2.55 | 1.75 | 5.2 | 0 | 0.5 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/12 RBS-lysis2-DT | 0.5 | 6.75 | 1.75 | 1.0 | 0 | 0.5 | 10.5
|
| genome DNA | Big Dye | MilliQ | 5x buffer | temp(400mg) | SasA_fwd primer | SasA_rev primer | total
|
| 9/11 RBS-lysis1-DT | 0.5 | 6.05 | 1.75 | 1.7 | 0 | 0.5 | 10.5
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 96°C | 96°C | 50°C | 68°C | --
|
| 2m | 10s | 5s | 4m | 30
|
PCR
Yoshida
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | Prefix-RBS-RpaB_fwd primer | RpaB-GG4-suffix_rev primer | MilliQ | total
|
| RpaB | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.5 | 25
|
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | Prefix-PkaiBC_fwd primer ver.3 | PkaiBC-suffix_rev primer ver.3 | MilliQ | total
|
| RpaB | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 58°C | 68°C | --
|
| 2m | 10s | 30s | 30s | 30
|
Nakamoto
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 3 | RBS-lacZα-DT | XbaI&PstI
|
| 4 | RBS-lacZα-DT | XbaI&PstI
|
| 5 | RBS-lacZα-DT | XbaI&PstI
|
| 6 | RBS-lacZα-DT | XbaI&PstI
|
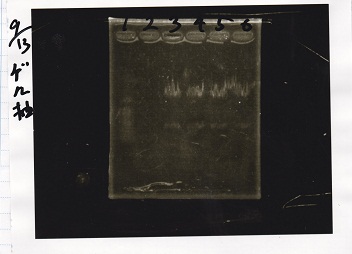

Transformation
No name
| Name | Sample | Competent Cells | Total | Plate
|
| 9/13 Plac+pT181 antisense | 2µL | 20µL | 22µL | Amp
|
| 9/13 Plac+apt12_1R | 2µL | 20µL | 22µL | Amp
|
| 9/13 Plac+pT181 attenuator | 2µL | 20µL | 22µL | Amp
|
| 9/13 Plux+pT181 antisense | 2µL | 20µL | 22µL | CP
|
| 9/13 Plux+RBS-lysis2-DT | 2µL | 20µL | 22µL | CP
|
| 9/13 Plux+RBS-lysis2-DT | 2µL | 20µL | 22µL | CP
|
| 9/13 Pcon-RBS-lacR-DT+Plux-RBS-GFP-DT | 2µL | 20µL | 22µL | Amp
|
| 9/13 Pcon-pT181 antisense+apt12_1R-DT | 2µL | 20µL | 22µL | Amp
|
| 9/13 Ptet-pT181antisense+spinach-DT | 2µL | 20µL | 22µL | CP
|
| Pλ-luxI-1 | 2µL | 20µL | 22µL | Amp
|
Sep 14
Restriction Enzyme Digestion
Hirano
| | 9/14 pSB4K5S | EcoRI | XbaI | SpeI | PstI | BufferB | BufferD | BSA | MilliQ | total
|
| 2cuts | 10µL | 1µL | 0µL | 1µL | 0µL | 3µL | 0µL | 3µL | 12µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 0µL | 1µL | 7.5µL | 10µL
|
| | 9/13 Plac | XbaI | PstI | BufferH | BSA | MilliQ | total
|
| 1 cut | 9µL | 0µL | 1µL | 3µL | 3µL | 14µL | 30µL
|
| NC | 1µL | 0µL | 0µL | 1µL | 1µL | 7µL | 10µL
|
| | 9/14 PKaiBC | EcoRI | XbaI | SpeI | PstI | BufferB | BSA | MilliQ | total
|
| 2cuts | 11µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 11µL | 30µL
|
| NC | 4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 4µL | 10µL
|
| | 9/14 RpaB | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total
|
| 2cuts | 13µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 9µL | 30µL
|
| NC | 2µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 5µL | 10µL
|
| | 8/17 RBS-GFP-DT | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total
|
| 1cut | 9µL | 0µL | 1µL | 0µL | 0µL | 3µL | 3µL | 14µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
Liquid Culture
Hirano
| Sample | medium
|
| 9/11 Plac(BBa-R0011)-(3) | Plusgrow medium(+Amp)
|
Electrophoresis
Hirano
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kb ladder | -- | --
|
| 2 | pSB4K5(1) | EcoRI | SpeI
|
| 3 | pSB4K5(1) | -- | --
|
| 4 | Plac(1A2) | PstI | --
|
| 5 | Plac(1A2) | -- | --
|
| 6 | PKaiBC | EcoRI | SpeI
|
| 7 | PKaiBC | -- | --
|
| 8 | RpaB(1) | XbaI | PstI
|
| 9 | RpaB(1) | -- | --
|
| 10 | RBS-GFP-DT | XbaI | --
|
| 11 | RBS-GFP-DT | -- | --
|
| 12 | 1kb ladder | -- | --
|
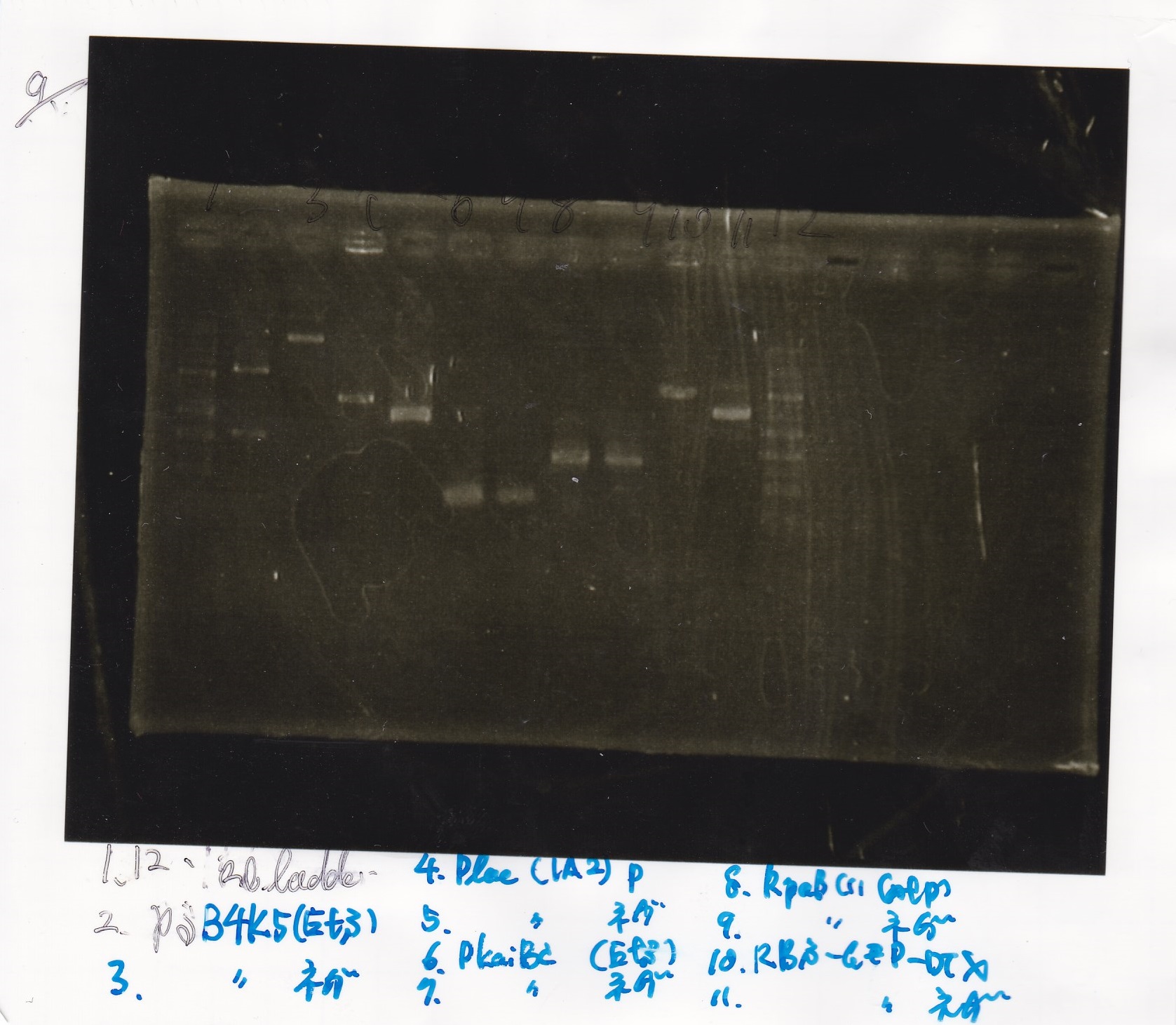
| Lane | Sample | Enzyme
|
| 1 | 1kb ladder | --
|
| 2 | Plac(pSB1A2) | PstI
|
| 3 | Plac(pSB1A2) | --
|

Sequence PCR
No name
| genome DNA | Big Dye | 5x buffer | temp(400mg) | primer | milliQ
|
| Pcon-RBS-luxR-DT | 0.5 | 1.75 | 0.9 | 0.5 | 6.85
|
| Pcon-RBS-luxR-DT | 0.5 | 1.75 | 0.4 | 0.5 | 7.35
|
| Pcon-RBS-tetR-DT | 0.5 | 1.75 | 3 | 0.5 | 5.25
|
| Pcon-RBS-tetR-DT | 0.5 | 1.75 | 3 | 0.5 | 5.25
|
| Plux | 0.5 | 1.75 | 2.4 | 0.5 | 5.35
|
| Plux-RBS-GFP-DT | 0.5 | 1.75 | 1.9 | 0.5 | 5.85
|
| Plux-RBS-GFP-DT | 0.5 | 0.5 | 1.9 | 0.5 | 5.85
|
| Pcon-RBS-GFP-DT | 0.5 | 0.5 | 1.2 | 0.5 | 6.55
|
| Pcon-RBS-GFP-DT | 0.5 | 0.5 | 1.2 | 0.5 | 6.55
|
| RBS-lysis3-DT | 0.5 | 0.5 | 1.0 | 0.5 | 6.75
|
| Pcon-antisense-Spinach-DT | 0.5 | 0.5 | 1.4 | 0.5 | 6.35
|
| Pcon-RBS lacZα-DT | 0.5 | 0.5 | 1.4 | 0.5 | 6.35
|
| Ptet-RBS lacZα-DT | 0.5 | 0.5 | 0.59 | 0.5 | 1.85
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 96°C | 96°C | 50°C | 60°C | --
|
| 2min | 10sec | 5sec | 4min | 30cycle
|
Tatsui
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 2 | pSB4K5 | EcoRI&SpeI
|
| 3 | pSB4K5 | EcoRI&SpeI
|
| 4 | pSB4K5 | EcoRI&SpeI
|


Restriction Enzyme Digestion
No name
| | 9/14 Plac(1A2) | EcoRI | XbaI | SpeI | PstI | BufferB | BSA | MilliQ | total
|
| 2cuts | 23µL | 0µL | 0µL | 0.3µL | 0µL | 3µL | 3µL | 0µL | 30µL
|
| | 9/14 RBS-GFP-DT | EcoRI | XbaI | SpeI | PstI | BufferH | BSA | MilliQ | total
|
| 2 cuts | 23µL | 1µL | 0µL | 0µL | 0µL | 3µL | 3µL | 0µL | 30µL
|
Ligation
Nakamoto and Hirano
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/13 Pcon-PT181attenuator(SpeI&PstI) | 1.7 | 9/13 RBS-lacZα-DT (XbaI & PstI) | 12.5 | 5
|
| experiment | 9/13 pSB2C3 (XbaI & PstI) | 1.9 | 9/14 RpaB (XbaI & PstI) | 16.3 | 10.5
|
| experiment | 9/13 pSB2C3 (XbaI & PstI) | 4.7 | 9/14 PkaiBC (EcoRI & SpeI) | 6.7 | 5.7
|
| experiment | 9/14 RBS-GFP-DT (PstI&XbaI) | 0.7 | 9/14 PkaiBC (EcoRI & SpeI) | 2.6 | 1.6
|
| experiment | 9/14 Plac (SpeI&PstI) | 2.4 | 9/14 RBS-lysis1-DT (XbaI & PstI) | 4.9 | 3.6
|
| experiment | 9/14 Plac (SpeI&PstI) | 2.4 | 9/14 RBS-lysis2-DT (XbaI & PstI) | 7.0 | 4.7
|
| experiment | 9/14 Plac (SpeI&PstI) | 2.4 | 9/14 RBS-lysis3-DT (XbaI & PstI) | 9.2 | 5.8
|
Samples were evaporeted used evaporator into about 7 µL
Colony PCR
No name
| Sample | base pair
|
| 9/13 Plux+RBS-lysis1-DT (1)~(3) | --
|
| 9/13 Plux+RBS-lysis2-DT (1)~(3) | -
|
| 9/13 Plux+RBS-lysis3-DT (1)~(2) | -
|
| Sample | base pair
|
| 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT (1)~(4) | --
|
| 9/13 Pcon-PT181attenuator+aptamer121R-DT (1)~(2) | -
|
| 9/13 Plac+PT181attenuator (1) | -
|
| Sample | base pair
|
| Dλ-lux(1) (1)~(2) | --
|
| NC | --
|
Electrophoresis
No name
| Lane | Sample
|
| 1 | 1kbp ladder
|
| 2 | 9/13 Plux+RBS-lysis1-DT1
|
| 3 | 9/13 Plux+RBS-lysis1-DT2
|
| 4 | 9/13 Plux+RBS-lysis1-DT3
|
| 5 | 9/13 Plux+RBS-lysis2-DT1
|
| 6 | 9/13 Plux+RBS-lysis2-DT2
|
| 7 | 9/13 Plux+RBS-lysis2-DT3
|
| 8 | 9/13 Plux+RBS-lysis3-DT1
|
| 9 | 9/13 Plux+RBS-lysis3-DT2
|
| 10 | 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT1
|
| 11 | 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT2
|
| 12 | 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT3
|
| 13 | 9/13 Pcon-RBS-luxR-DT+Plux-RBS-GFP-DT4
|
| 14 | 9/13 Ptet-pT181 antisense+Spinach-DT1
|
| 15 | 9/13 Pcon-pT181 attenuator+aptamer12-1R-PT1
|
| 16 | 9/13 Pcon-pT181 attenuator+aptamer12-1R-PT1
|
| 17 | 9/13 Plac+pT181 attenuator
|
| Lane | Sample
|
| 1 | 1kbp ladder
|
| 2 | Pλ-lux(1)1
|
| 3 | Pλ-lux(1)1
|
| 4 | NC
|

Liquid Culture
No name
| Sample | medium
|
| 9/13 Pλ-luxI(1)-1 | Plusgrow medium(+Amp)
|
Transformation
Nakamoto
| Name | Sample | Competent Cells | Total | Plate
|
| 9/13 RBS-lysisZα-DT(XbaI&PstI)+9/13 Pcon-PT181 attenuator(SpeI&PstI) | 2µL | 20µL | 22µL | Amp
|
| 9/13 SB2C3+9/14 PzαB | 2µL | 20µL | 22µL | CP
|
| 9/13 pSB2C3+9/14 PkaiBC | 2µL | 20µL | 22µL | CP
|
| 9/13 RBS-GFP-DT+9/14 PkaiBC | 2µL | 20µL | 22µL | CP
|
| Plac+RBS-lysis1-DT | 2µL | 20µL | 22µL | Amp
|
| Plac+RBS-lysis2-DT | 2µL | 20µL | 22µL | Amp
|
| Plac+RBS-lysis2-DT | 2µL | 20µL | 22µL | Amp
|
Sequence PCR
Tatsui
| genome DNA | temp | Big Dye | 5x buffer | primer | MilliQ
|
| 9/6 PT181 attenuator(VR) | 3.9 | 0.5 | 1.75 | 0.5 | 3.85
|
| 8/17 RBS lacZα-DT | 2.1 | 0.5 | 1.75 | 0.5 | 5.65
|
| 9/6 PT181 attenuator(VR) | 3.9 | 0.5 | 1.75 | 0.5 | 3.85
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 96°C | 50°C | 60°C | 4°C | --
|
| 10sec | 5sec | 4min | 1min | 30cycle
|
Sep 15
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| Pλ-luxI | 225 | 1.59 | 0.83
|
Liquid Culture
No name
| Sample | medium
|
| 9/13 Pλ-luxI-1 |
|
| 9/7 pSB4K5-1 |
|
| 9/11 Plac-3 |
|
Ligation
No name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/13 Pcon(SpeI & PstI) | 3.6 µL | 9/12 RBS-lacZα-DT(XbaI & PstI) | 4.7 µL | 8 µL
|
Restriction Enzyme Digestion
No Name
| | DNA | EcorI | SpeI | Buffer | BSA | MilliQ | total
|
| 9/7Pcon-RBS-tetR-DT (EcoRI & SpeI) | 15 µL | 1 µL | 1 µL | 3 µL | 3 µL | 6 µL | 30 µL
|
| NC | 0.7 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.3 µL | 10 µL
|
| | DNA | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 9/12 Pcon-pT181 attenuator-DT (EcoRI & SpeI) | 6.8 µL | 1 µL | 1 µL | 3 µL | 3 µL | 15.2 µL | 30 µL
|
| NC | 0.3 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.7 µL | 10 µL
|
| | DNA | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 9/12 Pcon-apt12_1R-DT(EcoRI & SpeI) | 6.6 µL | 1 µL | 1 µL | 3 µL | 3 µL | 15.4 µL | 30 µL
|
| NC | 0.3 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.7 µL | 10 µL
|
| | DNA | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 9/8 Pcon-spinach-DT (EcoRI & SpeI) | 7.6 µL | 1 µL | 1 µL | 3 µL | 3 µL | 15.4 µL | 30 µL
|
| NC | 0.3 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.7 µL | 10 µL
|
| | DNA | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 9/11 Pcon-pT181 anticodone-spinach-DT(XbaI & PstI) | 7.2 µL | 1 µL | 1 µL | 3 µL | 3 µL | 14.8 µL | 30 µL
|
| NC | 0.4 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.6 µL | 10 µL
|
| | DNA | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 8/17 RBS-lacZα-DT (XbaI & PstI) | 5.3 µL | 1 µL | 1 µL | 3 µL | 3 µL | 16.7 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.5 µL | 10 µL
|
| | DNA | EcoRI | XbaI | Buffer | BSA | MilliQ | total
|
| 9/7 Ptet-RBS-LacZα-DT (EcoRI & XbaI) | 2.9 µL | 1 µL | 1 µL | 3 µL | 3 µL | 18.5 µL | 30 µL
|
| NC | 1.5 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7 µL | 10 µL
|
Electrophoresis
No Name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1,14 | 100bp ladder | |
|
| 2 | Pcon-RBS-tetR-DT | EcoRI | SpeI
|
| 3 | Pcon-RBS-tetR-DT | |
|
| 4 | Pcon-pT181 attenuator-DT | EcoRI | SpeI
|
| 5 | Pcon-pT181 attenuator-DT | |
|
| 6 | Pcon-apt12-1R-DT | EcoRI | SpeI
|
| 7 | Pcon-apt12-1R-DT | |
|
| 8 | Pcon-spinach-DT | EcoRI | SpeI
|
| 9 | Pcon-spinach-DT | |
|
| 10 | Pcon-pT181 antisense-spinach-DT | XbaI | PstI
|
| 11 | Pcon-pT181 antisense-spinach-DT | |
|
| 12 | RBS-lacZα-DT | XbaI | PstI
|
| 13 | RBS-lacZα-DT | |
|
| 15 | Ptet-RBS-lacZα-DT | EcoRI | XbaI
|
| 16 | Ptet-RBS-lacZα-DT | |
|
| 17 | 1kbp ladder | |
|

No Name
1
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 2~4 | Pcon-pT181 attenuator-DT | EcoRI & SpeI
|
| 6~8 | Pcon-apt12_1R-DT |
|
| 10~12 | Pcon-spinach-DT |
|
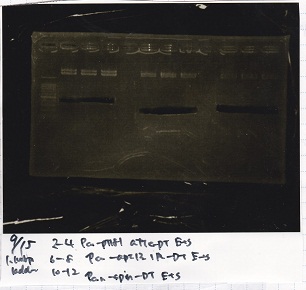
2
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder |
|
| 2~4 | Pcon-pT181 anticodone-spinach-DT | XbaI & PstI
|
| 6~8 | RBS-lacZα-DT | XbaI & PstI
|
| 10~12 | Pcon-RBS-tetR-DT | EcoRI & SpeI
|
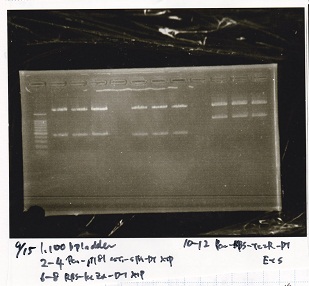

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 9/15 Ptet-RBS-lacZα-DT (EcoRI & SpeI) | 1.9 | 1.08 | 0.77
|
| 9/15 Pcon-RBS-tetR-DT (EcoRI & SpeI) | 5.9 | 1.30 | 0.17
|
| 9/15 RBS-lacZα-DT (XbaI & PstI) | 1.0 | 0.85 | 0.44
|
| Pcon-pT181 anticodone-spinach-DT (XbaI & PstI) | 1.3 | 0.95 | 0.28
|
| Pcon-spinach-DT | 0.9 | 1.37 | 0.05
|
| Pcon-apt12_1R-DT | 1.7 | 1.42 | 0.29
|
| Pcon-pT181 attenuator-DT (EcoRI & SpeI) | 2.1 | 1.53 | 0.46
|
Ligation
No Name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/13 pSB1C3 EcoRI & SpeI | 4.7 µL | 9/15 Pcon-RBS-tetR-DT EcoRI & SpeI | 1.9 µL | xx µL
|
| experiment | 9/13 pSB1C3 EcoRI & SpeI | 4.7 µL | 9/15 Pcon-spinach-DT EcoRI & SpeI | 4.7 µL | xx µL
|
| experiment | 9/13 pSB1C3 EcoRI & SpeI | 4.7 µL | 9/15 Pcon-apt12_1R-DT EcoRI & SpeI | 2.0 µL | xx µL
|
| experiment | 9/13 pSB1C3 EcoRI & SpeI | 4.7 µL | 9/15 Pcon-pT181 attenuator-DT EcoRI & SpeI | 3.0 µL | xx µL
|
| experiment | 9/13 pSB1C3 XbaI & PstI | 1.9 µL | 9/15 Pcon-pT181 anticodone-spinach-DT XbaI & PstI | 4.4 µL | xx µL
|
| experiment | 9/13 pSB1C3 EcoRI & SpeI | 4.7 µL | 9/3 pT181 attenuator EcoRI & SpeI | 3.8 µL | xx µL
|
| experiment | 9/6 Pcon SpeI & PstI | 1.4 µL | 9/15 RBS-lacZα-DT XbaI & PstI | 2.0 µL | xx µL
|
| experiment | 9/14 pSB 4k5 EcoRI & SpeI | µL | 9/15 Pcon-spinach-DT EcoRI & SpeI | 9.4 µL | xx µL
|
| experiment | 9/15 Peet-RBS-lacZα-DT EcoRI & XbaI | 2.6 µL | Pcon-RBS-tetR-DT EcoRI & SpeI | 1.5 µL | xx µL
|
Restriction Enzyme Digestion
No Name
| | DNA | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 9/13 pT181 attenuator XbaI & PstI | 6.3 µL | 1 µL | 1 µL | 3 µL | 3 µL | 15.7 µL | 30 µL
|
| NC | 0.3 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.7 µL | 10 µL
|
| | DNA | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 9/10 pSB1C3 EcoRI & SpeI | 8.1 µL | 1 µL | 1 µL | 3 µL | 3 µL | 13.9 µL | 30 µL
|
| NC | 0.4 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.6 µL | 10 µL
|
| | DNA | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 9/14 pSB4k5 XbaI & PstI | 9.5 µL | 1 µL | 1 µL | 3 µL | 3 µL | 12.5 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.5 µL | 10 µL
|
| | DNA | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 9/11 DT SpeI & PstI | 10.4 µL | 1 µL | 1 µL | 3 µL | 3 µL | 11.6 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.5 µL | 10 µL
|
| | DNA | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 9/6 apt12_1R-DT SpeI & PstI | 7.9 µL | 1 µL | 1 µL | 3 µL | 3 µL | 14.1 µL | 30 µL
|
| NC | 0.4 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.6 µL | 10 µL
|
| | DNA | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 9/10 spinach-DT SpeI & PstI | 9.9 µL | 1 µL | 1 µL | 3 µL | 3 µL | 12.1 µL | 30 µL
|
| NC | 0.5 µL | 0 µL | 0 µL | 1 µL | 1 µL | 7.5 µL | 10 µL
|
Liquid Culture
No name
| Sample | medium
|
| pSB1C3 |
|
| DT |
|
| Spinach-DT |
|
Sep 16
Liquid Culture
No name
| Sample | medium
|
| pSB1C3+P-KaiBC① | Plusgrow medium(+CP)
|
| pSB1C3+P-KaiBC② | Plusgrow medium(+CP)
|
Colony PCR
No name
| Sample | base pair
|
| pSB1C3+P-KaiBC① | 283
|
| pSB1C3+P-KaiBC② | 283
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 2min | 30s | 30s | 18s | 30cycles
|
No name
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder |
|
| 2 | pT181 attenuator | XbaI+PstI
|
| 3 | pT181 attenuator | XbaI+PstI
|
| 4 | pT181 attenuator | XbaI+PstI
|
| 6 | pSB1C3 | EcoRI+SpeI
|
| 7 | pSB1C3 | EcoRI+SpeI
|
| 8 | pSB1C3 | EcoRI+SpeI
|
| 10 | pSB4K5 | XbaI+PstI
|
| 11 | pSB4K5 | XbaI+PstI
|
| 12 | pSB4K5 | XbaI+PstI
|


| Name | concentration[µg/mL] | 260/280 | 260/230
|
| pT181 attenuator(XbaI+PstI) | 8.2 | 1.62 | 0.11
|
| pSB1C3(EcoRI+SpeI) | 49.5 | 1.48 | 0.18
|
| pSB4K5(XbaI+PstI) | 18.1 | 1.73 | 0.57
|
| DT(SpeI+PstI) | 70.4 | 1.36 | 1.02
|
| aptamer12-1R-DT(SpeI+PstI) | 42.0 | 1.82 | 0.47
|
| spinach-DT(SpeI+PstI) | 36.8 | 1.79 | 0.61
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| Dx-lux1 | 136.7 | 1.46 | 1.28
|
| RBS-laczα-DT | 237.6 | 1.88 | 1.70
|
| pSB4K5 | 148.4 | 1.55 | 1.09
|
| Pcon-KBS-tetR-DT | 126.6 | 1.73 | 1.36
|
| 9/11Plac1 | 160.0 | 1.78 | 1.28
|
| 9/15Plac2 | 144.0 | 1.73 | 1.81
|
| 9/14Plac | 332.5 | 1.51 | 2.33
|
| Ptet-RBS-laczα-DT | 231.3 | 1.84 | 1.57
|
PCR
No name
| DNA | 9/6pT181 antisense psB1C3(1) | KOD plus | 10x buffer | dNTP | MgSO4 | GG1_1C3_fwd primer | GG2_1C3_rev primer | MilliQ | total
|
| 400pg | 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| 800pg | 2 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 14.5 | 25
|
| 1000pg | 2.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 14 | 25
|
| nega | 0 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 30s | 30cycles
|
| DNA | 9/6 12-1R aptamer-DT | KOD plus | 10x buffer | dNTP | MgSO4 | GG1_1C3_fwd primer | GG1_1C3_rev primer | MilliQ | total
|
| 400pg | 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| 800pg | 2 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 14.5 | 25
|
| 1000pg | 2.5 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 14 | 25
|
| NC | 0 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 30s | 30cycles
|
Electrophoresis
Yoshida
| Lane | Sample
|
| 1 | 100bp ladder
|
| 2 | antisense 1000pg
|
| 3 | antisense 800pg
|
| 4 | antisense 400pg
|
| 5 | tet aptamer-DT 1000pg
|
| 6 | tet aptamer-DT 800pg
|
| 7 | tet aptamer-DT 400pg
|
| 8 | NC
|

Restriction Enzyme Digestion
Tatsui
| | 9/16 Pcon-RBS-tetR-DT(µL) | EcoRI(µL) | SpeI(µL) | XbaI(µL) | PstI(µL) | Buffer(µL) | BSA(µL) | MilliQ(µL) | total(µL)
|
| 2 cuts | 15.8 | 0 | 0 | 1 | 1 | 3 | 3 | 6.2 | 30
|
| NC | 0.8 | 0 | 0 | 0 | 0 | 1 | 1 | 7.2 | 10
|
| | 9/16 pSB4K5(µL) | EcoRI(µL) | SpeI(µL) | XbaI(µL) | PstI(µL) | Buffer(µL) | BSA(µL) | MilliQ(µL) | total(µL)
|
| 2 cuts | 13.5 | 0 | 0 | 1 | 1 | 3 | 3 | 8.5 | 30
|
| NC | 0.7 | 0 | 0 | 0 | 0 | 1 | 1 | 7.3 | 10
|
| | 8/22 pSB1C3(µL) | EcoRI(µL) | SpeI(µL) | XbaI(µL) | PstI(µL) | Buffer(µL) | BSA(µL) | MilliQ(µL) | total(µL)
|
| 2 cuts | 18.5 | 1 | 0 | 0 | 1 | 3 | 3 | 3.5 | 30
|
| NC | 0.9 | 0 | 0 | 0 | 0 | 1 | 1 | 7.1 | 10
|
| | 8/22 pSB1C3(µL) | EcoRI(µL) | SpeI(µL) | XbaI(µL) | PstI(µL) | Buffer(µL) | BSA(µL) | MilliQ(µL) | total(µL)
|
| 2 cuts | 18.5 | 1 | 1 | 0 | 0 | 3 | 3 | 3.5 | 30
|
| NC | 0.9 | 0 | 0 | 0 | 0 | 1 | 1 | 7.1 | 10
|
| | 8/21 pT181 attenuator2(µL) | EcoRI(µL) | SpeI(µL) | XbaI(µL) | PstI(µL) | Buffer(µL) | BSA(µL) | MilliQ(µL) | total(µL)
|
| 2 cuts | 7.4 | 1 | 1 | 0 | 0 | 3 | 3 | 14.6 | 30
|
| NC | 0.4 | 0 | 0 | 0 | 0 | 1 | 1 | 7.6 | 10
|
Ligation
Tatsui
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | Pcon-pT181antisense-spinach-DT(XbaI+PstI) | 5.0µg/mL | pSB4K5(XbaI+PstI) | 18.1µg/mL | xx µg/mL
|
| experiment | pC181 attenuator(XbaI+PstI) | 8.2µg/mL | pSB1C3(XbaI+PstI) | 26.0µg/mL | xx µg/mL
|
Transformation
tatsui
| Name | Sample | Competent Cells | Total | Plate
|
| Pcon+RBS-laczα-DT | µL | µL | µL |
|
| Pcon-pT181 attenuator-DT+pSB1C3 | µL | µL | µL |
|
| pT181 attenuator(EcoRI&SpeI)+pSB1C3 | µL | µL | µL |
|
| Pcon-aptamer12_1R-DT(EcoRI&SpeI)+pSB1C3 | µL | µL | µL |
|
| Pcon-spinach-DT(EcoRI&SpeI)+pSB1C3 | µL | µL | µL |
|
| Pcon-RBS-tetR-DT(EcoRI&SpeI)+pSB1C3 | µL | µL | µL |
|
| Pcon-pT181 antisense-spinach-DT(XbaI&PstI)+pSB1C3 | µL | µL | µL |
|
Miniprep
tatsui
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/15 pSBiC3 | 111.7 | 1.79 | 1.36
|
| 9/15 spinach-DT | 113.9 | 1.57 | 1.38
|
| 9/15 DT | 77.0 | 2.05 | 1.92
|
PCR
tatsui
| 9/14 aptamer12-P(VF) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 1.5 | 0.5 | 1.75 | 0.5 | 5.75 | 10.5
|
| 9/14 aptamer12-1M(VF) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 1.7 | 0.5 | 1.75 | 0.5 | 6.05 | 10.5
|
| 9/14 Fusion1 attenuator(VF) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 2.3 | 0.5 | 1.75 | 0.5 | 5.45 | 10.5
|
| 9/14 Fusion1 antisense(VF) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 1.3 | 0.5 | 1.75 | 0.5 | 6.45 | 10.5
|
| 9/13 Fusion3m2 attenuator(VF) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 1.7 | 0.5 | 1.75 | 0.5 | 6.05 | 10.5
|
| 9/13 Fusion6 antisense(VF) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 1.6 | 0.5 | 1.75 | 0.5 | 6.15 | 10.5
|
| 9/12 plux(VR) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 2.4 | 0.5 | 1.75 | 0.5 | 5.35 | 10.5
|
| 9/9 Ptet antisense(VF) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 2.2 | 0.5 | 1.75 | 0.5 | 5.55 | 10.5
|
| 9/12 Pcon-pT181 attenuator-DT(VF) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 1.4 | 0.5 | 1.75 | 0.5 | 6.35 | 10.5
|
| 9/12 Pcon-pT181 attenuator-DT(VR) | Big Dye | 5×Buffer | primer | MilliQ | total
|
| 1.4 | 0.5 | 1.75 | 0.5 | 6.35 | 10.5
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 96°C | 96°C | 50°C | 60°C | --
|
| 2min | 10s | 5s | 4min | 30cycles
|
Sep 17
Liquid Culture
Tatsui and Hirano
| Sample | medium
|
| Spinach-DT | LB medium(+CP)
|
| Pcon-Spinach-DT | LB medium(+Amp)
|
| Pcon-pT181 antisense-Spinach-DT | LB medium(+Amp)
|
PCR
Hirano
| Pcon-tetR aptamer-DT(VF) | BigDye | 5x buffer | primer(VF) | MilliQ | total
|
| 1.3 | 0.5 | 1.75 | 0.5 | 0.45 | 10.5
|
| Pcon-tetR aptamer-DT(VR) | BigDye | 5x buffer | primer(VR) | MilliQ | total
|
| 1.3 | 0.5 | 1.75 | 0.5 | 0.45 | 10.5
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 96 °C | 96 °C | 50 °C | 60 °C | --
|
| 2 min | 10 sec | 5 sec | 4 min | 30 cycles
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kbp ladder | -- | --
|
| 2 | Pcon-RBS-tetR-DT | XbaI | PstI
|
| 3 | Pcon-RBS-tetR-DT | -- | --
|
| 4 | pSB4K5 | XbaI | PstI
|
| 5 | pSB4K5 | -- | --
|
| 6 | pSB1C3 | EcoRI | SpeI
|
| 7 | pSB1C3 | EcoRI | PstI
|
| 8 | pSB1C3 | -- | --
|
| 9 | pT181 attenuator | EcoRI | SpeI
|
| 10 | pT181 attenuator | -- | --
|
| 11 | 1kbp ladder | -- | --
|
Ligation
No Name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 8/28 Plux (SpeI & PstI) | 5.0 µL | 9/13 RBS-lysis1-DT (XbaI & PstI) | 3.5 µL | 3.5 µL
|
| experiment | 8/28 Plux (SpeI & PstI) | 5.0 µL | 9/13 RBS-lysis2-DT (XbaI & PstI) | 3.6 µL | 3.5 µL
|
| experiment | 8/28 Plux (SpeI & PstI) | 11.0 µL | 9/13 RBS-lysis3-DT (XbaI & PstI) | 9.5 µL | 3.5 µL
|
| experiment | 9/14 Plac (SpeI & PstI) | 2.4 µL | 9/16 pT181 attenuator-DT (XbaI & PstI) | 4.8 µL | 3.5 µL
|
| experiment | 9/14 Plac (SpeI & PstI) | 2.4 µL | 9/3 pT181 antisense (XbaI & PstI) | 2.0 µL | 2.2 µL
|
| experiment | 9/14 Plac (SpeI & PstI) | 2.4 µL | 9/10 aptamer 12_1R-DT (XbaI & PstI) | 1.8 µL | 2.1 µL
|
| experiment | 9/13 pSB1C3 (XbaI & PstI) | 2.0 µL | 9/15 Pcon-PT181 antisense-Spinach-DT (XbaI & PstI) | 12.0 µL | 3.5 µL
|
| experiment | 9/16 pSB1C3 (EcoRI & SpeI) | 1.0 µL | 9/15 Pcon-Spinach-DT (EcoRI & SpeI) | 4.1 µL | 2.5 µL
|
| experiment | 9/16 pSB1C3 (EcoRI & SpeI) | 1.0 µL | 9/15 Pcon-RBS-tetR-DT (EcoRI & SpeI) | 2.0 µL | 1.5 µL
|
| experiment | 9/16 pSB1C3 (EcoRI & SpeI) | 1.0 µL | 9/15 Pcon-PT181 attenuator-DT (EcoRI & SpeI) | 2.4 µL | 1.7 µL
|
| experiment | 9/16 pSB1C3 (EcoRI & SpeI) | 1.0 µL | 9/15 Pcon-aptamer 12_1R-DT (EcoRI & SpeI) | 2.0 µL | 1.5 µL
|
| experiment | 9/13 pSB1C3 (XbaI & PstI) | 2.0 µL | 9/16 Pcon-pT181-attenuator (XbaI & PstI) | 4.4 µL | 3.2 µL
|
Mutation PCR
Kinose
| pC KaiABC(100pg/µL) | Prime STAR(2x) | Primer(KaiA-BsaI MuT-Fwd) | Primer(KaiA-BsaI MuT-Rev) | MilliQ | total
|
| 1.0 | 25.0 | 1.0 | 1.0 | 22.0 | 50.0
|
| 1.5 | 25.0 | 1.0 | 1.0 | 21.5 | 50.0
|
| 2.0 | 25.0 | 1.0 | 1.0 | 21.0 | 50.0
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 96 °C | 98 °C | 65 °C | 72 °C | --
|
| 5 min | 10 s | 15 s | 40 s | 30 cycles
|
Sep 18
Restriction Enzyme Digestion
No name
| | 9/18Plux | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 3.2µL | 1µL | 1µL | 3µL | 3µL | 18.8µL | 30µL
|
| NC | 1.7µL | 0µL | 0µL | 1µL | 1µL | 6.3µL | 10µL
|
| 9/18Pcon-pT181antisense-spinach-DT② | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 7.5µL | 1µL | 1µL | 3µL | 3µL | 14.5µL | 30µL
|
| NC | 1.4icro;L | 0µL | 0µL | 1µL | 1µL | 6.6µL | 10µL
|
| | 9/16 Plac② | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 13.8µL | 1µL | 1µL | 3µL | 3µL | 14.5µL | 30µL
|
| NC | 0.7µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL
|
| | 9/16 Spinach-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 17.6µL | 1µL | 1µL | 3µL | 3µL | 4.4µL | 30µL
|
| NC | 0.9µL | 0µL | 0µL | 1µL | 1µL | 7.1µL | 10µL
|
| | 9/8 Pcon J23100 | SpeI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 5.7µL | 1µL | 1µL | 3µL | 3µL | 16.3µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 9/16 RBS-lacZα-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 8.4µL | 1µL | 1µL | 3µL | 3µL | 13.6µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 9/11 Pcon-pT181antisense-spinach-DT①: | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 7.2µL | 1µL | 1µL | 3µL | 3µL | 14.8µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 9/6 apt12-1R-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 7.9µL | 1µL | 1µL | 3µL | 3µL | 14.1µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 9/3 pT181antisense | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 6.3µL | 1µL | 1µL | 3µL | 3µL | 15.7µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 1µL | 7.7µL | 10µL
|
| | 9/14 pSB4K5 | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 2cuts | 7.3µL | 1µL | 1µL | 3µL | 3µL | 14.7µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 8/31 Plux-RBS-GFP-DT | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 2cuts | 9.2µL | 1µL | 1µL | 3µL | 3µL | 12.8µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL
|
| | 9/12 RBS-lysis3-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts | 5.2µL | 1µL | 1µL | 3µL | 3µL | 16.8µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 9/18 Pcon-pT181attenuator | EcoRI | SpeI | Buffer | BSA | MilliQ | total
|
| 2cuts | 14.3µL | 1µL | 1µL | 3µL | 3µL | 7.7µL | 30µL
|
| NC | 0.7µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | Plae② | SpeI | PstI
|
| 2 | RBS-lysis3-DT | XbaI | PstI
|
| 3 | Plux-RBS-GFP-DT | EcoRI | SpeI
|
| 4 | PSB4K5 | EcoRI | SpeI
|
| 5 | Pcon | SpeI | PstI
|
| 6 | Plux | SpeI | PstI
|
| 7 | 1kbp ladder | -- | --
|
| 8 | Plae② | -- | --
|
| 9 | RBS-lysis3-DT | -- | --
|
| 10 | Plux-RBS-GFP-DT | -- | --
|
| 11 | PSB4K5 | -- | --
|
| 12 | Pcon | -- | --
|
| 13 | Plux | -- | --
|

Electrophoresis
Okazaki
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Pcon pT181antisense spinach-DT② | XbaI | PstI
|
| 3 | Spinach-DT | XbaI | PstI
|
| 4 | RBS-lacZα-DT | XbaI | PstI
|
| 5 | Pcon pT181antisense-Spinach-DT① | XbaI | PstI
|
| 6 | aptamer12-1R-DT | XbaI | PstI
|
| 7 | pT181attenuator | XbaI | PstI
|
| 8 | Pcon attenuator | EcoRI | SpeI
|
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Pcon pT181antisense spinach-DT② | XbaI | PstI
|
| 3 | Spinach-DT | XbaI | PstI
|
| 4 | RBS-lacZα-DT | XbaI | PstI
|
| 5 | Pcon pT181antisense-Spinach-DT① | XbaI | PstI
|
| 6 | apt12-1R-DT | XbaI | PstI
|
| 7 | pT181attenuator | XbaI | PstI
|
| 8 | Pcon attenuator | EcoRI | SpeI
|
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Pcon pT181antisense spinach-DT② | -- | --
|
| 3 | Spinach-DT | -- | --
|
| 4 | RBS-lacZα-DT | -- | --
|
| 5 | Pcon pT181antisense-Spinach-DT① | -- | --
|
| 6 | apt12-1R-DT | -- | --
|
| 7 | pT181attenuator | -- | --
|
| 8 | Pcon attenuator | -- | --
|
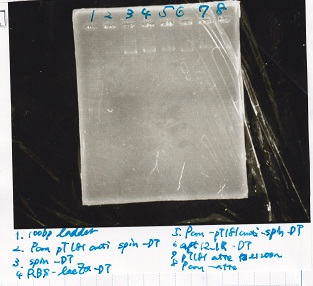


Sep 19
Sep 20
Ligation
Okazaki, Kanaoka, Tatsui and Hirano
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/18 Plac (SpeI & PstI) | 3.0 µL | 9/18 RBS-lysis3-DT (XbaI & PstI) | 12.4 µL | 3.5 µL
|
| experiment | 9/12 Ptet-pT181 antisense | 1.0 µL | 9/18 spinach-DT | 11.9 µL | 3.5 µL
|
| experiment | 9/13 Pcon-RBS-luxR-DT | 2.1 µL | 9/10 Plux-RBS-GFP-DT | 5.6 µL | 3.5 µL
|
| experiment | 9/13 Pcon-pT181 attenuator (SpeI & PstI) | 2.0 µL | 9/18 aptamer 12-1R-DT (XbaI & PstI) | 6.3 µL | 3.5 µL
|
| experiment | 9/18 Plac ②(SpeI & PstI) | 3.0 µL | 9/18 aptamer 12-1R-DT(XbaI & PstI) | 4.5µL | x3.5 µL
|
Transformation
Tatsui
| Name | Sample | Competent Cells | Plate
|
| 9/17 pSB1C3-Pcon-pT181 attenuator-DT | 2 µL | 20 µL | CP
|
| 9/17 pSB1C3-Pcon-aptamer 12_1R-DT | 2 µL | 20 µL | CP
|
| 9/17 pSB1C3-Pcon-antisense-spinach-DT | 2 µL | 20 µL | CP
|
| 9/17 pSB1C3-Pcon-RBS-tetR-DT x2 | 2 µL | 20 µL | CP
|
| 9/14 pSB1C3-PkaiBC | 2 µL | 20 µL | CP
|
| 9/14 pSB1C3-RpaB | 2 µL | 20 µL | CP
|
| 9/16 pSB1C3-pT181 attenuator(EcoRI&SpeI) | 2 µL | 20 µL | CP
|
| 9/17 pSB1C3-pT181 attenuator(XbaI&PstI) | 2 µL | 20 µL | CP
|
| 9/17 pSB1C3-Pcon-spinach-DT | 2 µL | 20 µL | CP
|
| 9/14 RBS-GFP-DT-PkaiBC | 2 µL | 20 µL | CP
|
| Plux-RBS-lysis3-DT | 2 µL | 20 µL | CP
|
| 9/20 Ptet-pT181 antisense-spinach-DT | 2 µL | 20 µL | CP
|
| 9/20 Pcon-spinach-DT(pSB4K5) | 2 µL | 20 µL | Kan
|
| 9/20 Pcon-pT181 antisense-spinach-DT(pSB4K5) | 2 µL | 20 µL | Kan
|
| 9/20 KaiABC | 2 µL | 20 µL | Kan
|
| 9/20 KaiABC | 2 µL | 20 µL | Kan
|
| 9/20 Plac-RBS-lysis3-DT | 2 µL | 20 µL | Amp
|
| 9/20 Plac-aptamer 12_1R | 2 µL | 20 µL | Amp
|
| 9/20 Plux-RBS-GFP-DT-Pcon-RBS-luxR-DT | 2 µL | 20 µL | Amp
|
| Plac-aptamer12_1R-DT | 2 µL | 20 µL | Amp
|
| 9/20 Plac-aptamer12_1R-DT | 2 µL | 20 µL | Amp
|
| 9/20Pcon-pT181attenuator-aptamer-DT | 2 µL | 20 µL | Amp
|
| 9/17 Plac-pT181-attenuatorx2 | 2 µL | 20 µL | Amp
|
| 9/15 Pcon-RBS-lacZα-DT | 2 µL | 20 µL | Amp
|
| 9/14 Pcon-pT181attenuator-RBS-lacZα-DT | 2 µL | 20 µL | Amp
|
| PUC19 | 2 µL | 20 µL | Amp
|
PCR
Tatsui
| Sample | base pair
|
| pT181 attenuator(pSB1C3) | --
|
| Pcon-RBS-lacZα-DT | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 94 °C | 55 °C | 68 °C | --
|
| 5 min | 30 s | 30 s | 3 min | 30 cycles
|
Electrophoresis
No name
| Lane | Sample | Enzyme
|
| 1 | Pcon-RBS-lacZα-DT | --
|
| 2 | 1kb ladder | --
|
| 3 | pSB1C3-pT181attenuator | --
|
Sep 21
Electrophoresis
no name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | Pcon-RBS-lacZα-DT | -- | --
|
| 2 | 1kbp ladder | -- | --
|
| 3 | pSB1C3+pT181 attenuator | -- | --
|
Liquid Culture
No name
| Sample | medium
|
| 9/2 spinach-DT-2 | --
|
| 8/16 Plux-4 | --
|
| 8/27 Plux-RBS-GFP-DT-1 | --
|
| 9/5 Pcon-pT181 attenuator-1 | --
|
Master Plate
No name
| Number | Use LB plate
|
| 1 | 8/27 Pλ-luxI-1(+Amp)
|
| 2 | 9/10 Plac(BBa-R0011)-3(+Amp)
|
| 3 | 9/5 Pcon-RBS-tetR-DT-1(+Amp)
|
| 4 | 9/5 Pcon-pT181 antisense-1(+Amp)
|
| 5 | 9/15 Pcon-pT181 attenuator(+Amp)
|
| 6 | 9/- Pcon-apt12_1R-DT-1(+Amp)
|
| 7 | 9/5 Pcon-spinach-DT-3(+Amp)
|
| 8 | 8/27 Pcon-RBS-GFP-DT-Pcon-RBS-luxR-DT(+Amp)
|
| Number | Use LB plate
|
| 1 | 9/4 pT181 antisense(pSB1C3)-1(+CP)
|
| 2 | 9/11 Fusion1 antisense(pSB1C3)-4(+CP)
|
| 3 | 9/11 Fusion6 antisense(pSB1C3)-1(+CP)
|
| 4 | 9/4 pT181 attenuator(pSB1C3)-1(+CP)
|
| 5 | 9/10 Fusion1 attenuator(pSB1C3)-1(+CP)
|
| 6 | 9/11 Fusion3n2 attenuator(pSB1C3)-1(+CP)
|
| 7 | 9/4 aptamer12_1R(pSB1C3)-1(+CP)
|
| 8 | 9/11 aptamer12_P(pSB1C3)-3(+CP)
|
| 9 | 9/11 aptamer12_1M(pSB1C3)-1(+CP)
|
| Number | Use LB plate
|
| 1 | 8/29 Pbad/araC-RBS-RFP-DT-1(+CP)
|
| 2 | 9/4 RBS-lysis3-DT-3(+CP)
|
| 3 | 9/10 Pcon-pT181 attenuator-DT-1(+CP)
|
| 4 | 9/5 Ptet-pT181 antisense-1(+CP)
|
| 5 | 9/4aptamer 12_1R-DT-1(+CP)
|
| 6 | 9/5 Ptet-RBS-lacZα-DT-4(+CP)
|
| 7 | 8/10 RBS-lacI-DT-2(+CP)
|
| 8 | 8/29 Plux-RBS-GFP-DT-1(+CP)
|
Restriction Enzyme Digestion
Noname
| | 8/17 RBS-DT | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts(XbaI&PstI) | 8.1 | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 13.9µL | 30µL
|
| NC | 0.4 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| | 8/17 RBS-DT | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cut(EcoRI&XbaI) | 16.2 | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 5.8µL | 30µL
|
| | 9/15 Pcon-pT181attenuator | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cuts(EcoRI&PstI) | 12 | 1µL | 0µL | 0µL | 1µL | 3µL | 3µL | 10µL | 30µL
|
| NC | 0.6 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.4µL | 10µL
|
| | 8/20 Pcon=RBS-GFP-DT-1 | EcoRI | SpeI | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2cut(EcoRI&XbaI) | 6 | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 0µL | 14µL
|
| NC | 0.3 | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL
|
incubate 37°C 1hour
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | RBS-GFP-DT | XbaI | PstI
|
| 2 | RBS-GFP-DT | -- | --
|
| 3 | RBS-GFP-DT | EcoRI | XbaI
|
| 4 | 1kbp ladder | -- | --
|
| 5 | Pcon-pT181attenuator | SpeI | PstI
|
| 6 | Pcon-pT181attenuator | -- | --
|
| 7 | Pcon-RBs-GFP-DT | EcoRI | XbaI
|
| 8 | Pcon-RBs-GFP-DT | -- | --
|

PCR
Nakamoto
| Sample | base pair
|
| Pcon-apt12_1R-DT(E-1A) | 50
|
| Pcon-spinach-DT(E-1A) | 402
|
| Pcon-pT181 attenuator(E-1A) | 400
|
| DT(1-3) | 65
|
| tetR aptamer 12_1R-DT(1-3) | 283
|
| Pcon-pT181 attenuator(1-SA) | 303
|
| spinach-DT(2-1) | 338
|
| Pcon-pT181 attenuator(E-0) | 403
|
| Pcon-pT181 attenuator(2-SA) | 403
|
| spinach-DT(3-2) | 395
|
| Ptet-PT181 anisense | 400
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 35s | 30cycles
|
No name
| Lane | DNA | Enzyme
|
| 1 | 1kb ladder | --
|
| 3 | RBS-GFP-DT | XbaI&PstI
|
| 4 | RBS-GFP-DT | XbaI&PstI
|
| 5 | RBS-GFP-DT | XbaI&PstI
|


| Name | concentration[µg/mL] | 260/280 | 260/230
|
| RBS-GFP-DT(XbaI&PstI) | 14.8 | 1.74 | 0.47
|
| RBS-GFP-DT(EcoRI&SpeI) | 88.4 | 1.85 | 1.49
|
| Pcon-RBS-GFP-DT(EcoRI&XbaI) | 47.5 | 1.82 | 1.12
|
Sep 22
Colony PCR
Tatsui
| Sample | base pair
|
| 9/20 Plac-PT181 attenuator(1~4) | 664
|
| 9/20 Plac-PT181 antisense(1~6) | 369
|
| 9/20 Pcon-PT181 attenuator-aptamer-DT(1~6) | 859
|
| 9/20 Pcon-PT181 antisense-spinach-DT(1~8) | 723
|
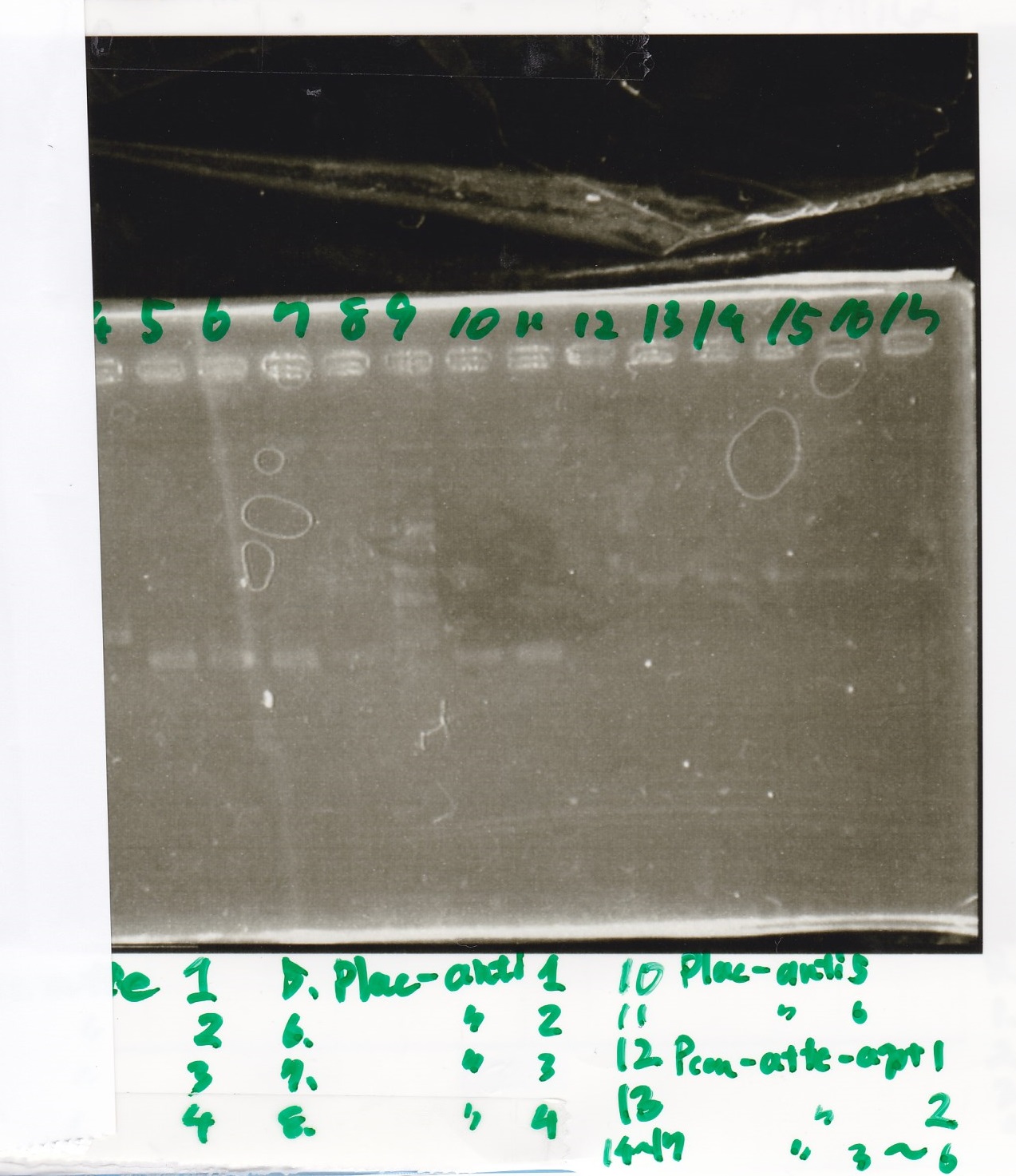
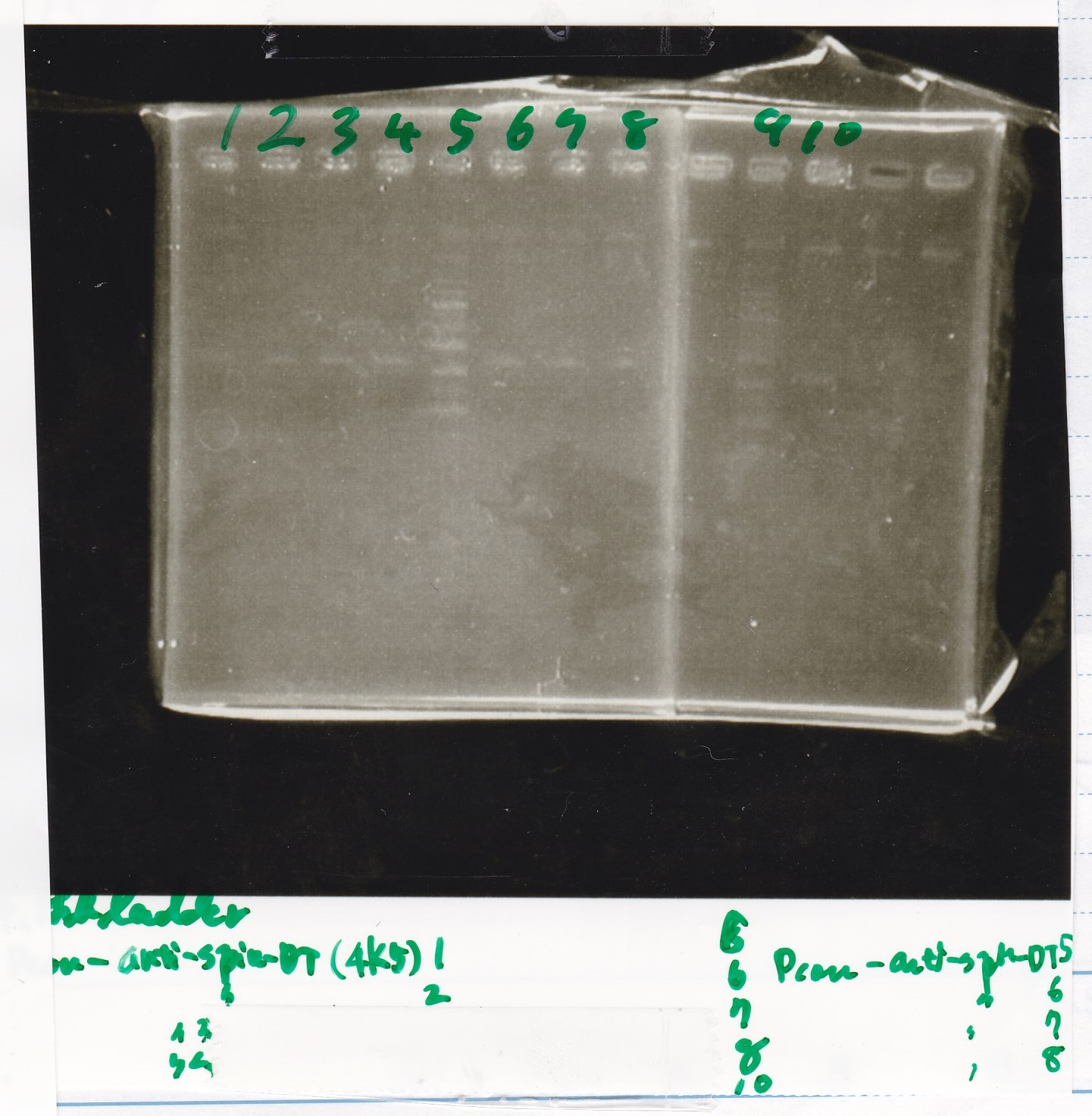
Electrophoresis
Nakamoto
| Lane | Sample
|
| 1 | Pcon-apt12_1R-DT(E-1A)
|
| 2 | Pcon-spinach-DT(E-1A)
|
| 3 | Pcon-pT181 attenuator(E-1A)
|
| 4 | DT(1-3)
|
| 5 | tetR aptamer 12_1R-DT(1-3)
|
| 6 | 100bp ladder
|
| 7 | Pcon-pT181 attenuator(1-SA)
|
| 8 | spinach-DT(2-1)
|
| 9 | Pcon-pT181 attenuator(E-0)
|
| 10 | Pcon-pT181 attenuator(2-SA)
|
| 11 | spinach-DT(3-2)
|
| 12 | Ptet-PT181 anisense
|


Miniprep
Honda
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/21 spinach-DT | 197.9 | 1.67 | 1.54
|
| 9/21 Plux | 125.9 | 1.71 | 1.27
|
| 9/21 Plux-RBS-GFP-DT-(1) | 168.0 | 1.62 | 1.73
|
| 9/21 Pcon-PT181 attenuator-(1) | 368.3 | 1.57 | 1.62
|
| 9/21 RBS-GFP-DT | 532.8 | 1.75 | 1.78
|
| 9/21 Pcon-RBS-GFP-DT | 845.2 | 2.02 | 1.70
|
Colony PCR
Honda
| Sample | base pair
|
| 9/20 Plux-RBS-GFP-DT-Pcon-RBS-luxR-DT-(1~2) | 283
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 2min10s | 30cycles
|
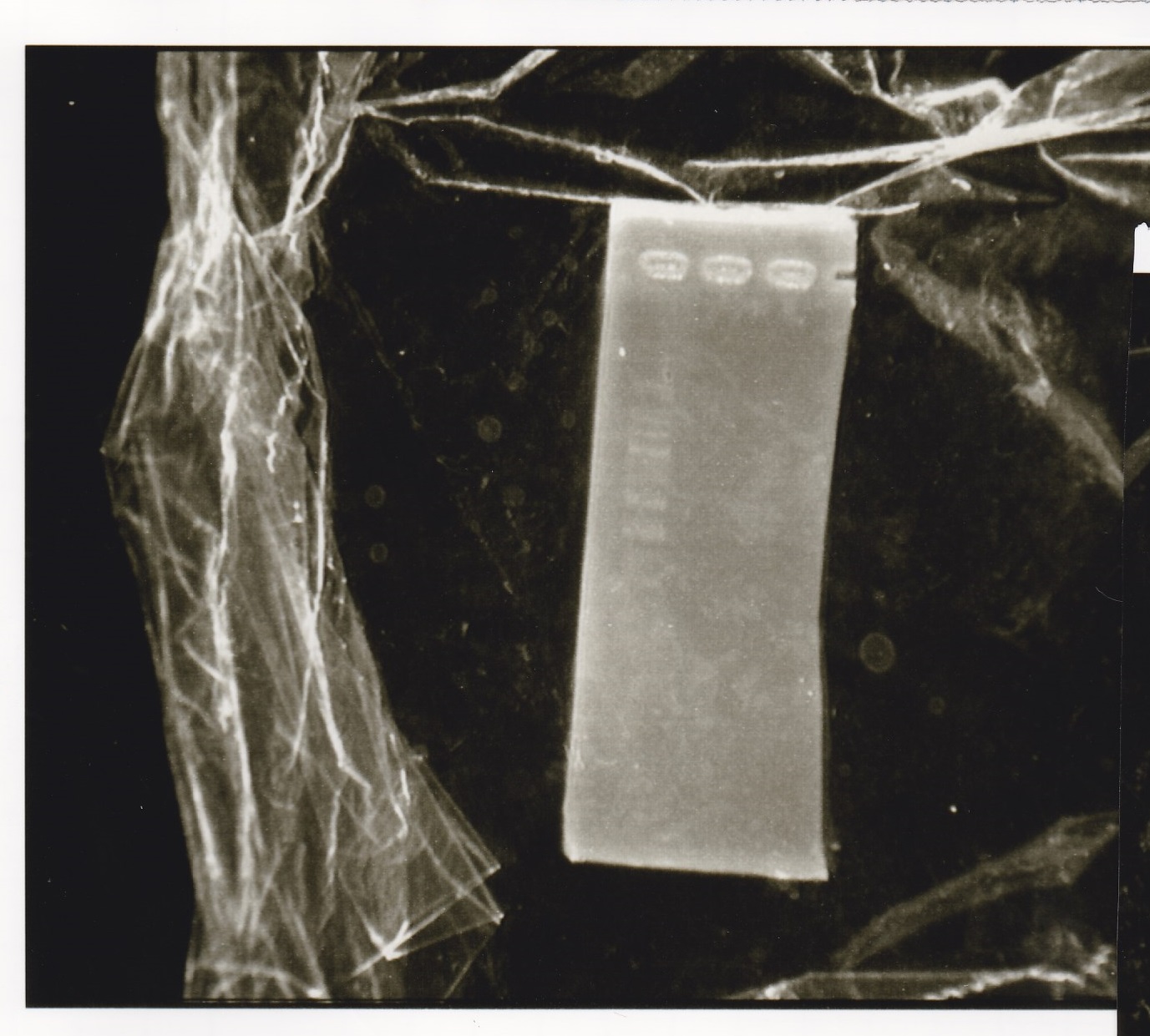

No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2~4 | Pcon-aptamer12-1R-DT(E-1A) | --
|
| 6~8 | Pcon-spinach-DT(E-1A) | --
|
| 10~12 | Pcon-attenuator PT181(E-1A) | --
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2~4 | DT(1-3) | --
|
| 6~8 | aptamer12-1RDT(1-3) | --
|
| 10~12 | Pcon-PT181 attenuator(1-5A) | --
|
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-aptamer12-1R-DT(E-1A) | 31.2 | 1.87 | 0.30
|
| Pcon-spinach-DT(E-1A) | 25.6 | 1.88 | 0.99
|
| Pcon-attenuator PT181(E-1A) | 25.3 | 1.99 | 0.86
|
| DT(1-3) | 22.1 | 1.82 | 0.20
|
| aptamer12-1RDT(1-3) | 33.5 | 1.99 | 0.05
|
| Pcon-PT181 attenuator(1-5A) | 36.9 | 1.12 | 0.96
|
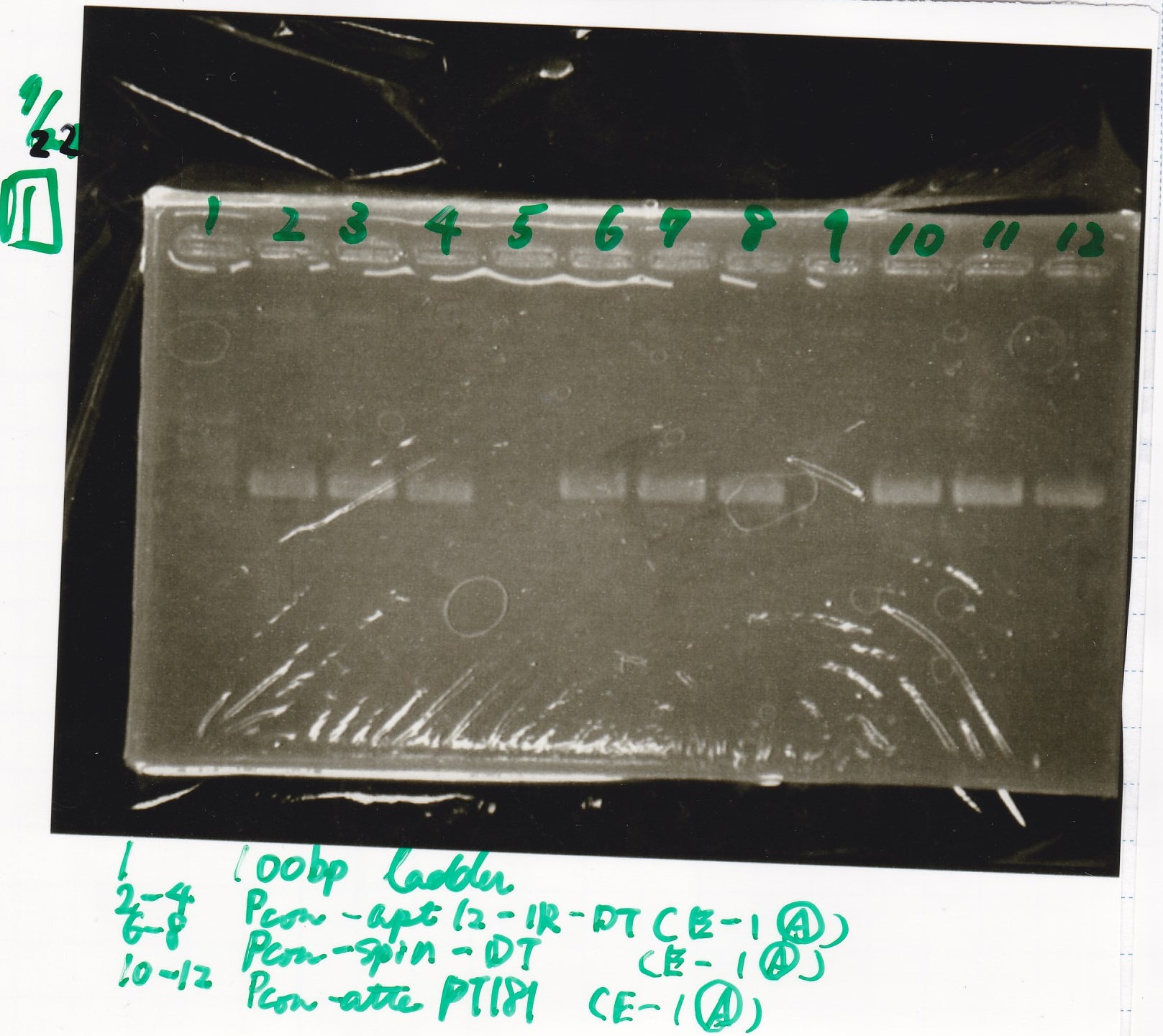



Restriction Enzyme Digestion
No name
| | 9/16 Plac | XbaI | SpeI | PstI | BufferB | BSA | MilliQ | total
|
| 2cuts | 12.5µL | 0µL | 1µL | 1µL | 3µL | 3µL | 9.5µL | 30µL
|
| NC | 0.8µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.4µL | 10µL
|
| | 9/22 Plux | XbaI | SpeI | PstI | BufferB | BSA | MilliQ | total
|
| 2cuts | 16µL | 0µL | 1µL | 1µL | 3µL | 3µL | 14µL | 30µL
|
| NC | 0.8µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.2µL | 10µL
|
| | 9/20 RBS-tetR-2R-DT | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total
|
| 2cuts | 8.5µL | 1µL | 0µL | 1µL | 3µL | 3µL | 13.5µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
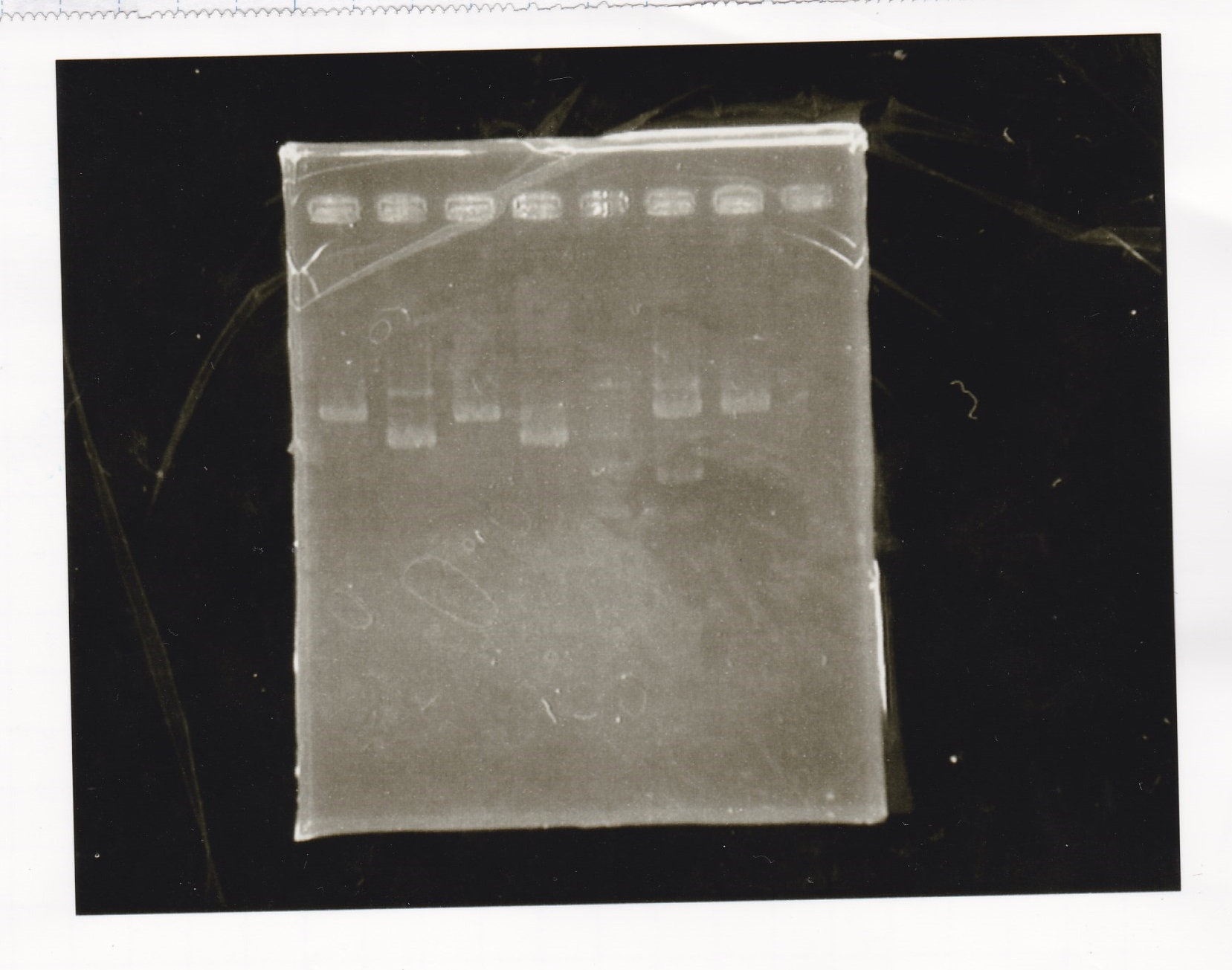
No name
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2~4 | Pcon-attenuator PT181(E-0) | --
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2~4 | Pcon-attenuator PT181(E-0) | --
|
| Lane | DNA | Enzyme
|
| 2 | 100bp ladder | --
|
| 4~6 | Pcon-attenuator PT181(2-SA) | --
|
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 3~5 | spinach-DT(3-2) | --
|
| 8~10 | Ptet-antisense PT181(1-3) | --
|
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-attenuator PT181(E-0) | 2.4 | 1.80 | 0.17
|
| Pcon-attenuator PT181(2-SA) | 10.8 | 1.85 | 0.54
|
| spinach-DT(3-2) | 36.7 | 1.80 | 0.75
|
| Ptet-antisense PT181(1-3) | 16.8 | 1.74 | 0.78
|

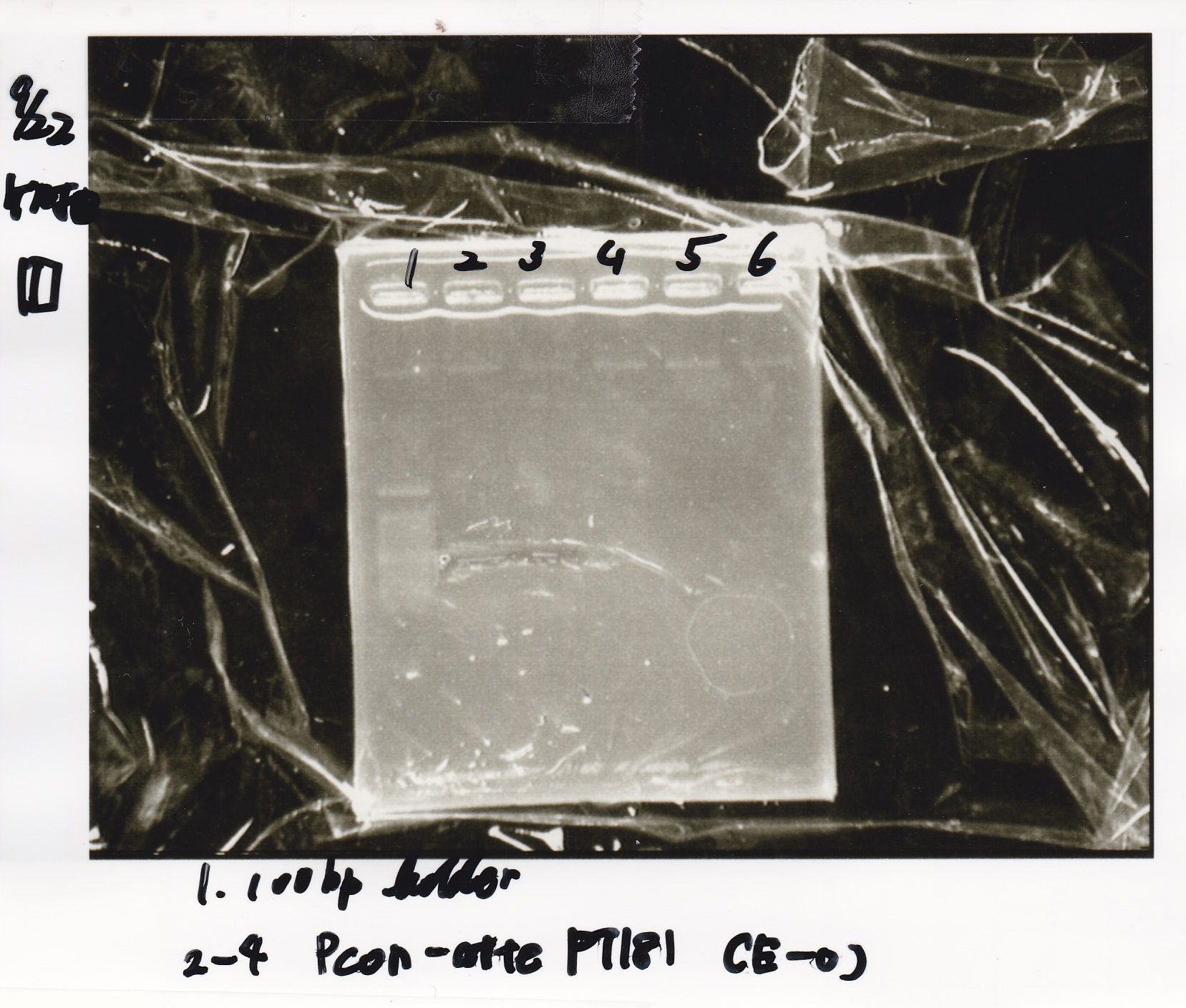




Colony PCR
No name
| Sample | base pair
|
| 9/20 Pcon-PT181 attenuator-aptamer12-1R-DT-(7~14) | 859
|
Colony PCR
Tatsui
| Sample | base pair
|
| PconPT181antisense-spinach-DT(9~15) | 723
|

Ligation
No name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/13 Pcon-PT181 attenuator (SpeI & PstI) | 1.7 µL | 9/18 aptamer-1R-DT (XbaI & PstI) | 5.4µL | 3.5µL
|
| experiment | 9/12 Plac(SpeI & PstI) | 2.4 µL | 9/18 aptamer-1R-DT (XbaI & PstI) | 6.1µL | 3.5 µL
|
| experiment | 9/13 Ptet(SpeI & PstI) | 1.7 µL | 9/10 Plux-RBS-GFP-DT | 7.0 µL | 4.4 µL
|
| experiment | 9/21 RBS-GFP-DT (EcoRI & XbaI) | 0.6 µL | 9/18 aptamer 12-1R-DT (XbaI & PstI) | 5.6µL | 3.1 µL
|
| experiment | 9/13 PSB1C3(XbaI & PstI) | 1.9 µL | 9/16 PT181 attenuator(XbaI & PstI) | 4.6µL | 3.3 µL
|
Liquid Culture
No name
| Sample | medium
|
| 9/20 Plac-antisense-1 | --
|
| 9/20 Plac-attenuator-1 | --
|
| 9/20 Plac-antisense-1 | --
|
| 9/20 Plac-attenuator-1 | --
|
digestion
No name
| | 9/8 Pcon-PT181 antisense | EcoRI | SpeI | BufferM | MilliQ | total
|
| 2cuts | 8.3µL | 1µL | 1µL | 3µL | 16.7µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 8.3µL | 10µL
|
| | 9/7 Pcon-RBS-tetR-DT | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total
|
| 2cut | 14µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 18.2µL | 30µL
|
| NC | 0.7µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL
|
| | 9/22 RBS-GFP-DT | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ | total
|
| 2cuts | 3.8µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 18.2µL | 30µL
|
| NC | 0.2µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.8µL | 10µL
|
| | 9/6 aptamer12-1R-DT | EcoRI | XbaI | SpeI | PstI | BufferD | BSA | MilliQ
|
| 2cut | 8µL | 1µL | 1µL | 0µL | 0µL | 3µL | 3µL | 16µL
|
| NC | 0.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL
|
incubate 37°C 1hour

No name
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 3~5 | Pcon-PT181 antisense (EcoRI&SpaI) | --
|
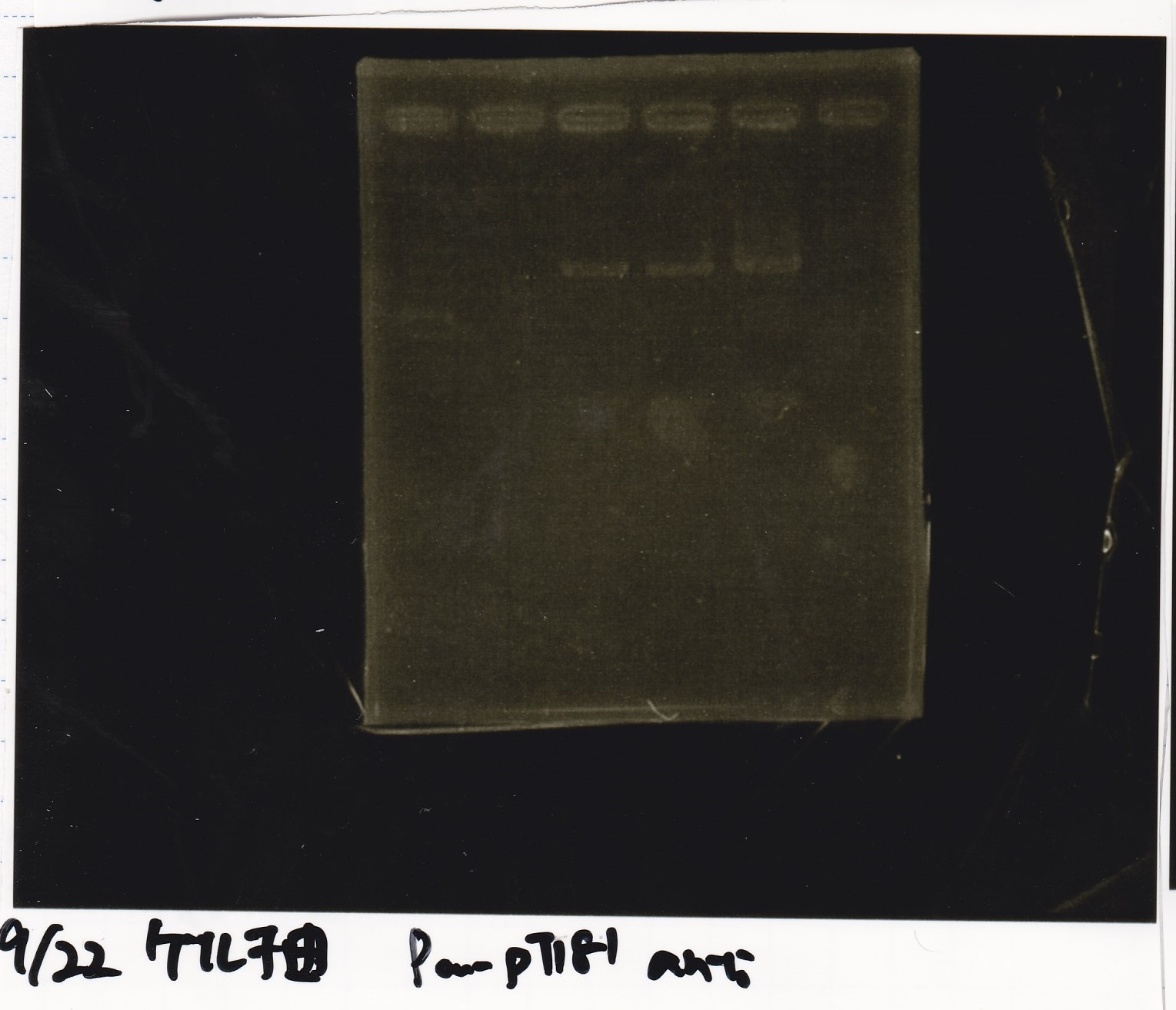
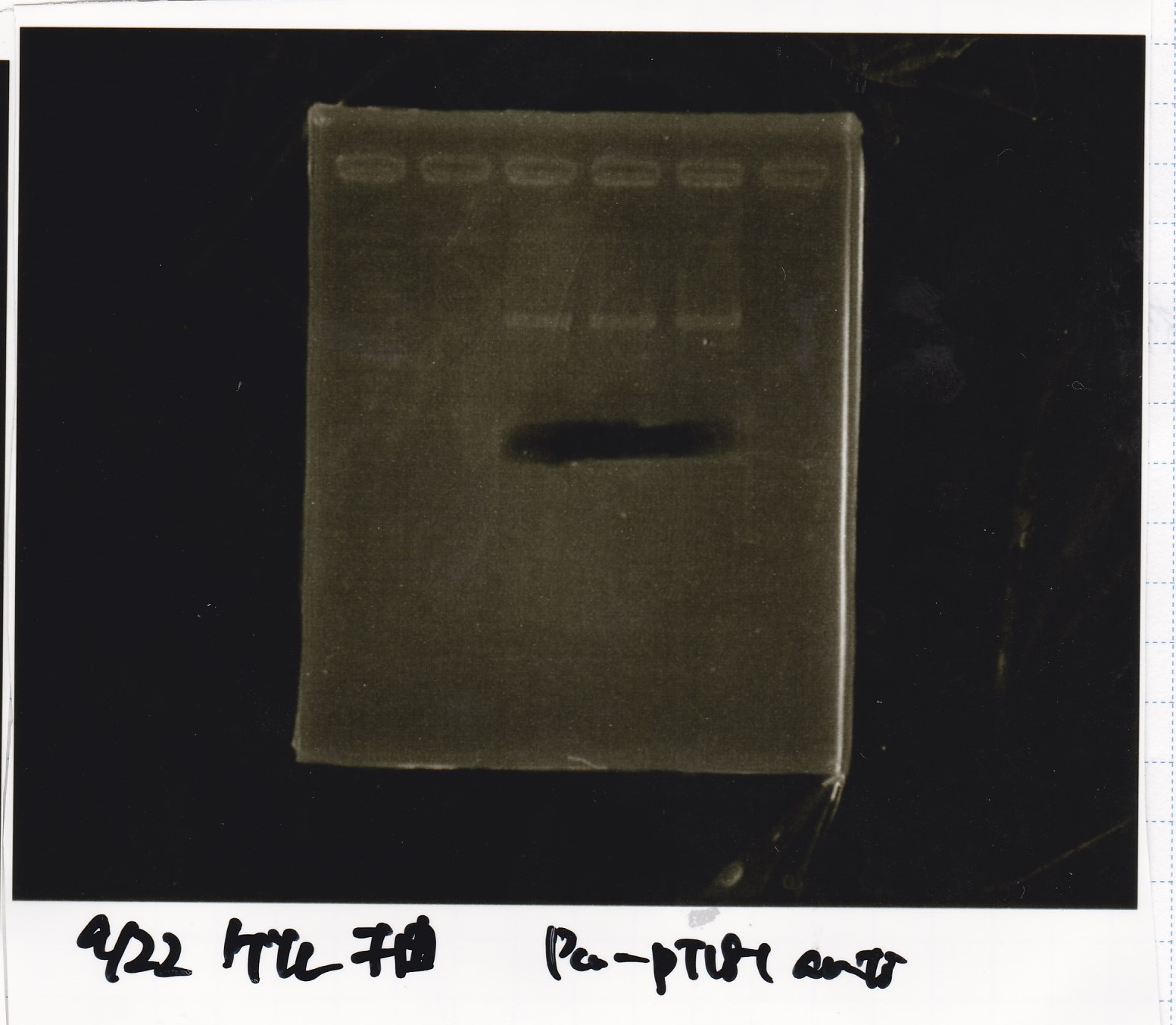
Transformation
No name
| Name | Sample | Competent Cells | Plate
|
| 9/20 Plac-aptamer12-1R-DT(XbaI&PstI) | 2 µL | 20 µL | Amp
|
| 9/20 pT181 attenuator(1C3)(EcoRI&SpeI) | 2 µL | 20 µL | CP
|
| 9/17 pT181 attenuator(1C3)(XbaI&PstI) | 2 µL | 20 µL | CP
|
| 9/20 Pcon-pT181 attenuator(SpaI&PstI)+aptamer12-1R-DT(XbaI&PstI) | 2 µL | 20 µL | Amp
|
| kaiABC | 2 µL | 20 µL | Amp
|
| 9/22 pSB1C3-RpaB(XbaI&PstI) | 2 µL | 20 µL | Amp
|
| 9/22 pSB1C3-pT181 attenuator(XbaI&PstI) | 2 µL | 20 µL | Amp
|
| 9/22 Ptet(SpaI&PstI)+RBS-GFP-DT(XbaI&PstI) | 2 µL | 20 µL | CP
|
| 9/22 RBS-GFP-DT+Pcon-attenuator pT181 (EcoRI&SpeI) | 2 µL | 20 µL | CP
|
| 9/22 pT181 attenuator(XbaI&PstI)(1C3) | 2 µL | 20 µL | CP
|
PCR
No name
| Sample | base pair
|
| Pcon-RBS-tetR-DT(1-2(A)) | --
|
| Pcon-RBS-tetR-DT(3-2(A)) | --
|
| Pcon-RBS-GFP-DT(1-S(A)) | --
|
| Pcon-RBS-GFP-DT(2-S(A)) | --
|
| Pcon-tetR-DT(E-2(A)) | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 94 °C | 57 °C | 68 °C | --
|
| 2 min | 10 s | 30 s | 1min5s | 30 cycles
|
Electrophoresis
No name
| Lane | Sample
|
| 1 | 1kbp ladder
|
| 2 | Pcon-RBS-tetR-DT(1-2Ⓐ)
|
| 3 | Pcon-RBS-tetR-DT(3-2Ⓐ)
|
| 4 | Pcon-RBS-GFP-DT(1-SⒶ)
|
| 5 | Pcon-RBS-GFP-DT(2-SⒶ)
|
| 6 | Pcon-tetR-DT(E-2Ⓐ)
|

Sep 23
Transformation
No name
| Name | Sample | Competent Cells | Plate
|
| 9/14 pSB1C3+PkaiBC | 2 µL | 20 µL | --
|
| 9/14 pSB1C3+RpaB | 2 µL | 20 µL | --
|
| 9/14 pkaiBC+Rbs-GFP-DT | 2 µL | 20 µL | --
|
No Name
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder |
|
| 3-4 | Pcon-RBS-tetR-PT |
|

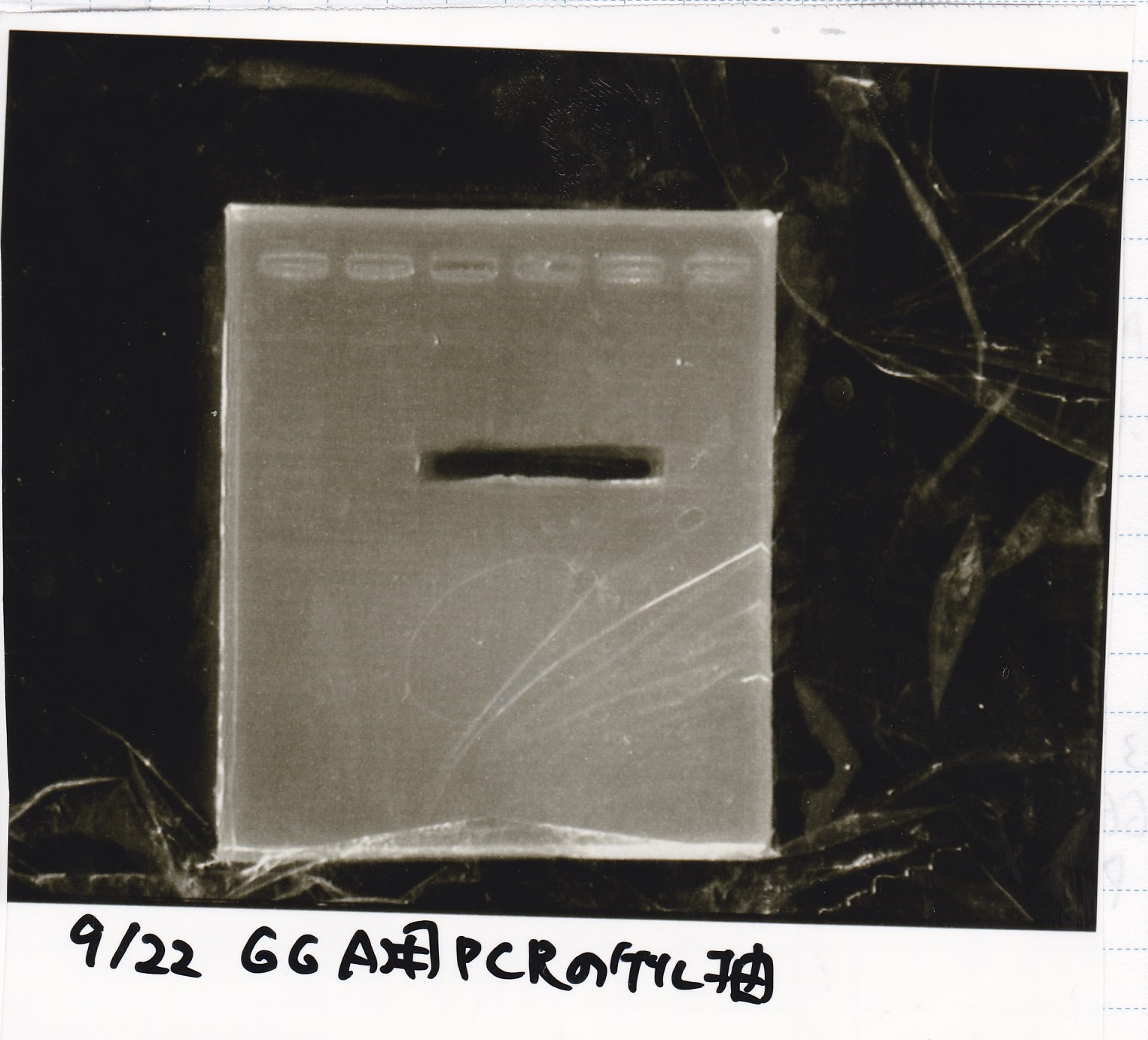
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-PT181 anti EcoRI+SpeI | 46.7 | 1.06 | 0.93
|
| Pcon-RBS-tetR-DT EcoRI+XbaI | 119.3 | 0.99 |
|
| RBS-8FP-DT EcoRI+XbaI | 87.7 | 1.00 | 0.98
|
| ap42_1R-DT EcoRI+XbaI | 68.8 | 1.32 | 1.03
|
| Pcon-RBS-tetR-DT 1-2 A | 203.0 | 1.66 | 1.79
|
| Pcon-RBS-tetR-DT 3-2 A | 67.7 | 2.64 | 1.28
|
| Pcon-RBS-GFP-DT 1-S A | 185.6 | 1.88 | 2.04
|
| Pcon-RBS-GFP-DT 2-S A | 298.6 | 1.68 | 1.55
|
| Pcon-tetR-DT E-2 A | 202.4 | 1.57 | 1.66
|
PCR
No name
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | SasA_fwd primer | SasA_rev primer | MilliQ | total
|
| 9/8 Pcon-pT181 anti | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| 8/20 Ptet(1) | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| 9/16 Plac 2 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 98 °C | 57 °C | 68 °C | --
|
| 2min | 10sec | 30sec | 20sec | 30cycles
|
Ligation
Yoshida
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/21 Pcon-RBS-GFP-DT EcoRI+XbaI | 47.5 µg/mL | 9/15 Pcon-RBS-tetR-DT EcoRI+SpeI | 59 µg/mL | 2.3 µg/mL
|
| experiment | 9/21 Pcon-RBS-GFP-DT EcoRI+XbaI | 47.5 µg/mL | 9/15 Pcon-apt12_1R-DT EcoRI+SpeI | 17 µg/mL | 2.4 µg/mL
|
| experiment | 9/21 Pcon-RBS-GFRDT EcoRI+XbaI | 2.1 µg/mL | 9/15 Pcon-DT181 attenuator-DT | 21 µg/mL | 3.0 µg/mL
|
| experiment | 9/22 apt 12_1R-DT EcoRI+XbaI | 67.8 µg/mL | 9/18 Pcon-DT181 attenuator EcoRI+SpeI | 5.7 µg/mL | 3.5 µg/mL
|
| experiment | 9/22RBS-GFP-DT EcoRI+XbaI | 87.7 µg/mL | 9/18 Pcon-DT18 attenuator EcoRI+SpeI | 6.7 µg/mL | 3.5 µg/mL
|
| experiment | 9/13 DT EcoRI+XbaI | 25.4 µg/mL | 9/22 Pcon-DT181 antisense EcoRI+SpeI | 46.8 µg/mL | 2.3 µg/mL
|
PCR
No name
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | SasA_fwd primer | SasA_rev primer | MilliQ | total
|
| Pcon-attenuator-DT (E-1A) | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 77.5 | 25
|
| Pcon-attenuator-DT (2-1A) | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 77.5 | 25
|
| Pcon-antisense-spinach-DT (1-2A) | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 77.5 | 25
|
| Pcon-antisense-spinach-DT (E-1A) | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 77.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 98 °C | 57 °C | 68 °C | --
|
| 2min | 10s | 30s | 45s | 30cycle
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/22 Plac-DT181 attenuator-1 | 204.0 | 2.00 | 1.95
|
| 9/22 Plac-DT181 attenuator-2 | 195.3 | 1.84 | 1.62
|
| 9/22 Plac-DT181 antisense-1 | 188.2 | 1.87 | 2.15
|
| 9/22 Plac-DT181 antisense-2 | 165.0 | 1.95 | 2.66
|
Electrophoresis
Hirano
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1,6 | 100bp ladder(Promega) | -- | --
|
| 2 | Pcon-attenuator-DT (E-1A) | -- | --
|
| 3 | Pcon-attenuator-DT (2-1A) | -- | --
|
| 4 | Pcon-antisense-spinach-DT (1-2A) | -- | --
|
| 5 | Pcon-antisense-spinach-DT (E-1A) | -- | --
|
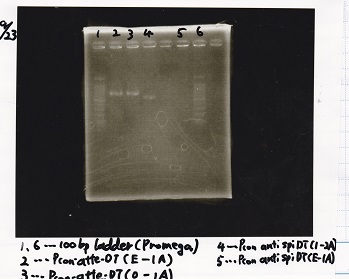
Hirano
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-pT181 attenuator-DT(E-1A) | 166.7 | 1.51 | 0.99
|
| Pcon-pT181 attenuator-DT(2-1A) | 122.1 | 1.75 | 1.20
|
| Pcon-pT181 antisense-spinach-DT(1-2A) | 99.1 | 1.86 | 1.81
|
Electrophoresis
Hirano
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | 9/8 Pcon-pT181 antisense(E-1A) | -- | --
|
| 3 | 8/20 Ptet(1)(2-S) | -- | --
|
| 4 | 9/16 Plac2(E-1A) | -- | --
|

Transformation
No Name
| Name | Sample | Competent Cells | Total | Plate
|
| pSB1C3 | 2µL | 20µL | 22µL |
|
Ligation
No Name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/19 Plac SpeI+PstI | 24.6 µg/mL | 9/18 ape12_1R-DT XbaI+PstI | 4.5 µg/mL | 2 µg/mL
|
| experiment | 9/21 RBS-GFP-DT EcoRI+XbaI | 88.4 µg/mL | 9/15 Pcon-pT181 attenuator EcoRI+SpeI | 21 µg/mL | 2 µg/mL
|
| experiment | 9/13 PSB1C3 XbaI+PstI | 26.0 µg/mL | 9/16 pT181 attenuator XbaI+PstI | 8.2 µg/mL | 2 µg/mL
|
Colony PCR
Tatsui
| Sample | base pair
|
| 9/22 Pcon-PT181 attenuator12-1R-DT(1~8) | --
|
| 9/22 Pcon-PT181 attenuator(1~4) | --
|
| 9/20 Plac+aptamer12-1R-DT(1~4) | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 54s | 30cycle
|
Sep 24
Electrophoresis
No name
| Lane | Sample
|
| 1 | 9/20 Pcon-pT181aptamer12-1R-DT 1
|
| 2 | 9/20 Pcon-pT181aptamer12-1R-DT 2
|
| 3 | 9/20 Pcon-pT181aptamer12-1R-DT 3
|
| 4 | 9/20 Pcon-pT181aptamer12-1R-DT 4
|
| 5 | 9/20 Pcon-pT181aptamer12-1R-DT 5
|
| 6 | 9/20 Pcon-pT181aptamer12-1R-DT 6
|
| 7 | 9/20 Pcon-pT181aptamer12-1R-DT 7
|
| 8 | 9/20 Pcon-pT181aptamer12-1R-DT 8
|
| 9 | 100bp ladder
|
| 10 | 9/22 Pcon-pT181attenuator-aptamer12-1R-DT 1
|
| 11 | 9/22 Pcon-pT181attenuator-aptamer12-1R-DT 2
|
| 12 | 9/22 Pcon-pT181attenuator-aptamer12-1R-DT 3
|
| 13 | 9/22 Pcon-pT181attenuator-aptamer12-1R-DT 4
|
| 14 | 9/20 Plac-aptamer12-1R-DT 1
|
| 15 | 9/20 Plac-aptamer12-1R-DT 2
|
| 16 | 9/20 Plac-aptamer12-1R-DT 3
|
| 17 | 9/20 Plac-aptamer12-1R-DT 4
|
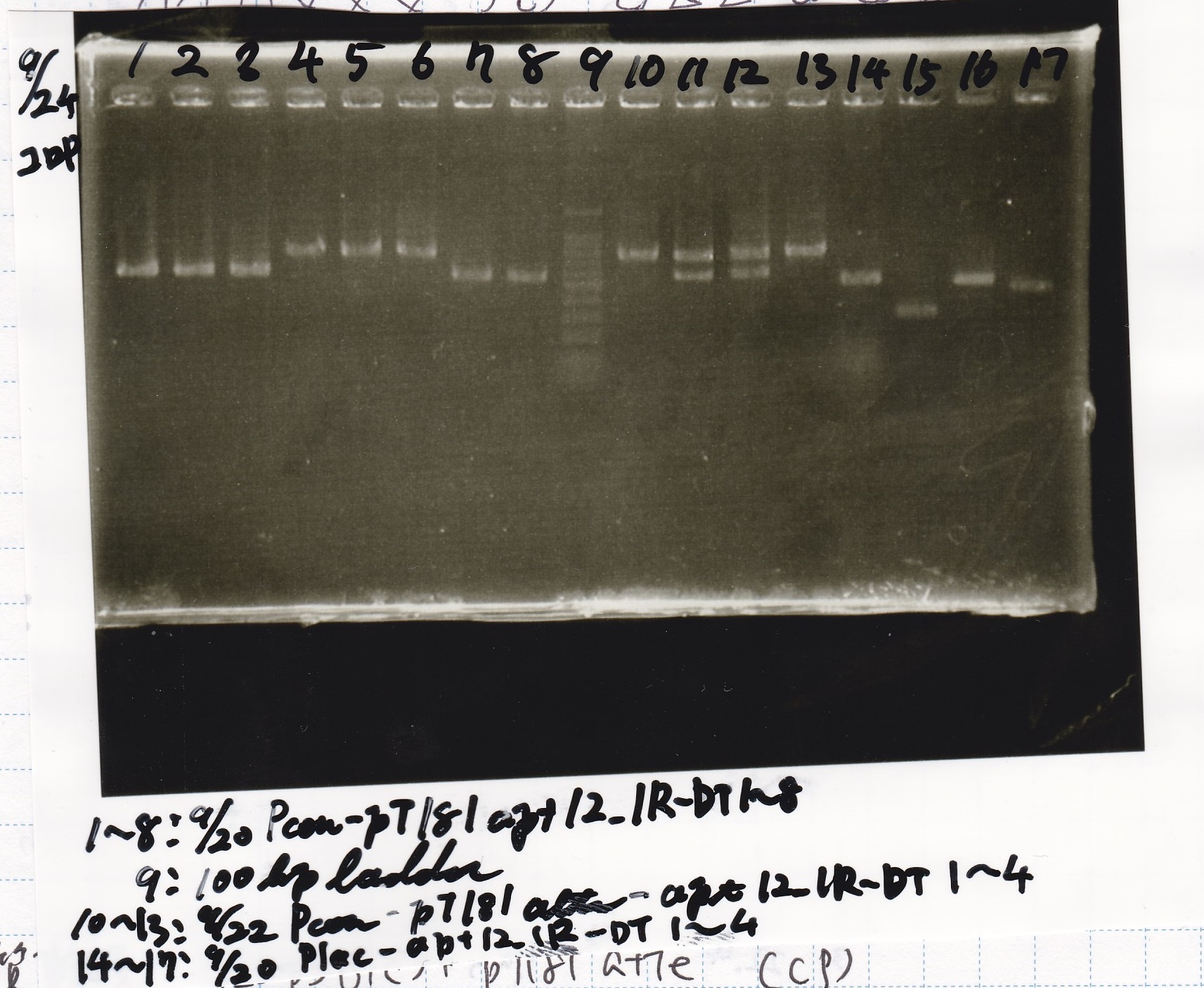
Transformation
No name
| Name | Sample | Competent Cells | Total | Plate
|
| 9/23 pSB1C3-pT181attenuator | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| RBS-GFP-DT+Pcon-pT181attenuator | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| 9/23 Plac-+spinach-DT | 2 µL | 20 µL | 22 µL | LB (+Amp)
|
| 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT | 2 µL | 20 µL | 22 µL | LB (+Amp)
|
| 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT | 2 µL | 20 µL | 22 µL | LB (+Amp)
|
| 9/23 Pcon-RBS-tetR-DT+Pcon-RBS-GFP-DT | 2 µL | 20 µL | 22 µL | LB (+Amp)
|
| 9/23 Pcon-pT181antisense-DT | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| 9/23 Pcon-pT181attenuator-RBS-GFP-DT | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| 9/23 Pcon-pT181attenuator-aptamer12-1R-DT | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| PUC19 | 2 µL | 20 µL | 22 µL | LB (+Amp)
|
| Ptet-RBS-lacZα-DT | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| pSB1C3 | 2 µL | 20 µL | 22 µL | LB (+CP)
|
Liquid Culture
No name
| Sample | medium
|
| aptamer12-1R-DT(Master plate) | LB(+CP)
|
Restriction Enzyme Digestion(PCR product)
Hirano
Concentrate PCR product by evaporator until volume become less than 1µL, add all sulution and pipetting.
| 10*cut smart buffer | BsaI-HF | MilliQ | total
|
| 9/22 Pcon-pT181 attenuator (E-0) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-pT181 attenuator (2-3A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 spinach-DT(3-2) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Ptet-pT181 antisense (1-3) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-tet aptamer-DT (E-1A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-spinach-DT (E-1A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-pT181 attenuator (E-1A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 DT (1-3) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 tet aptamer-DT (1-3) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-pT181 attenuator (1-SA) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-RBS-tetR-DT (1-2A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-RBS-tetR-DT (3-2A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-RBS-tetR-DT (E-2A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-RBS-GFP-DT (1-2A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/22 Pcon-RBS-GFP-DT (2-SA) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/23 Pcon-pT181 attenuator-DT (E-1A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/23 Pcon-pT181 attenuator-DT (0-1A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/23 Pcon-pT181 antisense-spinach-DT (1-2A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/23 Pcon-pT181 antisense (1-2A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/23 Ptet-1 (2-S) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
| 9/23 Plac-2 (E-1A) | 2.0 µL | 0.3 µL | 17 µL | 19.3µL
|
Incubate 37°C 12hour
Column Refining
Kojima
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/22 Pcon-pT181 attenuator(E-0) | 7.8 | 1.70 | 0.03
|
| 8/22 Pcon-pT181 attenuator(2-SA) | 17.3 | 1.93 | 0.03
|
| 8/22 spinach-DT (3-2) | 22.4 | 1.93 | 0.09
|
| 8/22 Ptet-PT181 anisense (1-3) | 18.3 | 1.58 | 0.14
|
| 8/22 Pcon-aptamer12_1R-DT (E-1A) | 8.2 | 2.10 | 0.05
|
| 8/22 Pcon-spinach-DT(E-1A) | 5.6 | 0.91 | 0.05
|
| 8/22 Pcon-pT181 attenuator(E-1A) | 18.5 | 1.80 | 0.12
|
| 8/22 DT(1-3) | 7.3 | 1.13 | 0.04
|
| 8/22 tetR aptamer 12_1R-DT(1-3) | 14.6 | 1.74 | 0.06
|
| 8/22 Pcon-pT181 attenuator(1-SA) | 7.9 | 1.57 | 0.04
|
| 8/22 Pcon-RBS-tetR-DT(1-2Ⓐ) | 41.6 | 1.81 | 0.25
|
| 8/22 Pcon-RBS-tetR-DT(3-2Ⓐ) | 30.2 | 1.75 | 0.15
|
| 8/22 Pcon-RBS-tetR-DT(E-2Ⓐ) | 36.9 | 1.75 | 0.56
|
| 8/22 Pcon-RBS-GFP-DT(1-SⒶ) | 43.5 | 1.83 | 0.33
|
| 8/22 Pcon-RBS-GFP-DT(2-SⒶ) | 64.6 | 1.82 | 0.30
|
| 8/23 Pcon-pT181 attenuator-DT (E-1A) | 31.2 | 1.83 | 0.09
|
| 8/23 Pcon-pT181 attenuator-DT (0-1A) | 64.5 | 1.80 | 0.43
|
| 8/23 Pcon-pT181 antisense-spinach-DT (1-2A) | 36.4 | 1.88 | 0.21
|
| 8/23 Pcon-pT181 antisense-spinach-DT (E-1A) | 27.9 | 1.92 | 0.16
|
| 8/23 Ptet(1)(2-S) | 37.0 | 1.91 | 0.16
|
| 9/23 Plac-2(E-1A) | 21.2 | 1.82 | 0.13
|
Transformation
No name
| Name | Sample | Competent Cells | Total | Plate
|
| 9/23 Pcon-attenuator-aptamer12-1R-DT | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| 9/23 pT181attenuator(1C3) | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| 9/23 Pcon-pT181antisense-DT | 2 µL | 20 µL | 22 µL | LB (+CP)
|
| 9/23 Pcon-pT181attenuator-RBS--GFP-DT | 2 µL | 20 µL | 22 µL | LB (+CP)
|
Liquid Culture
No name
| Sample | medium
|
| pSB1C3 (Master plate-1) | LB(+CP)
|
Restriction Enzyme Digestion
Kojima
| | 8/21 pT181attenuator (330 µg/mL) | XbaI | PstI | buffer | BSA | MilliQ | total
|
| 2 cuts | 6.1 | 1 | 1 | 3 | 3 | 15.9 | 30
|
| NC | 0.3 | 0 | 0 | 1 | 1 | 7.7 | 10
|
| | 9/22 Pcon-pT181attenuator (368.3 µg/mL) | EcoRI | SpeI | buffer | BSA | MilliQ | total
|
| 2 cuts | 5.4 | 1 | 1 | 3 | 0 | 19.6 | 30
|
| NC | 0.3 | 0 | 0 | 1 | 0 | 8.7 | 10
|
| | 9/16 pSB1C3 | EcoRI | SpeI | buffer | BSA | MilliQ | total
|
| 2 cuts | 18 | 1 | 1 | 3 | 0 | 7.0 | 30
|
| NC | 0.9 | 0 | 0 | 1 | 0 | 8.1 | 10
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kb ladder | - | -
|
| 2 | pT181attenuator | XbaI | PstI
|
| 3 | pT181attenuator | -- | --
|
| 4 | Pcon-pT181attenuator | EcoRI | SpeI
|
| 5 | Pcon-pT181attenuator | -- | --
|
| 6 | pSB1C3 | EcoRI | SpeI
|
| 7 | pSB1C3 | -- | --
|
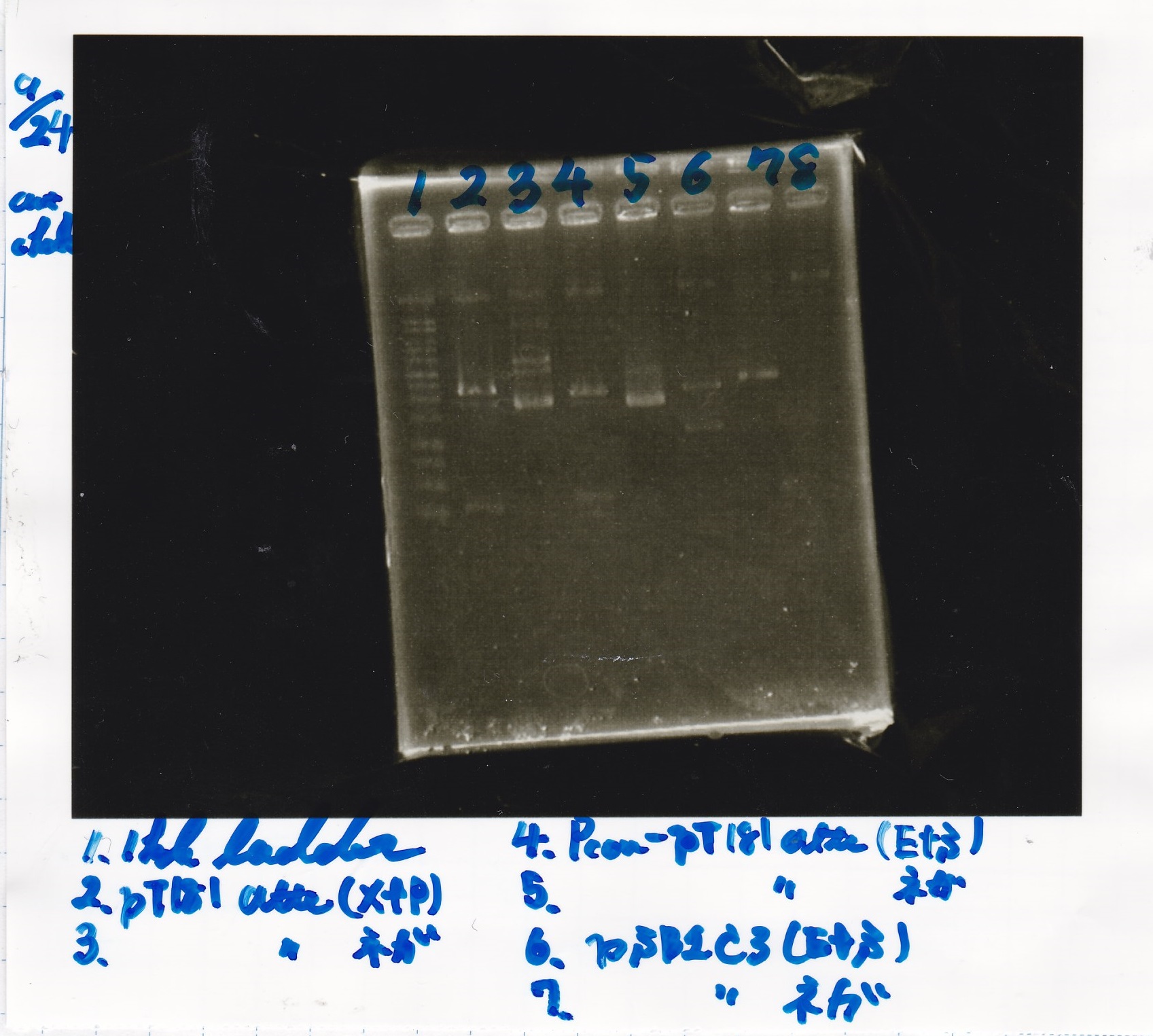
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| 9/24 aptamer12-1R-DT | 144.7 | 1.03 | 1.21
|
Kojima
| Lane | DNA | Enzyme1 | Enzyme2
|
| 1 | 100kb ladder | -- | --
|
| 2 | pT181attenuator | XbaI | PstI
|
| 3 | pT181attenuator | XbaI | PstI
|
| 4 | pT181attenuator | XbaI | PstI
|
| 5 | -- | -- | --
|
| 6 | Pcon-pT181attenuator | EcoRI | SpeI
|
| 7 | Pcon-pT181attenuator | EcoRI | SpeI
|
| 8 | Pcon-pT181attenuator | EcoRI | SpeI
|
| 9 | -- | -- | --
|
| 10 | pSB1C3 | EcoRI | SpeI
|
| 11 | pSB1C3 | EcoRI | SpeI
|
| 12 | pSB1C3 | EcoRI | SpeI
|
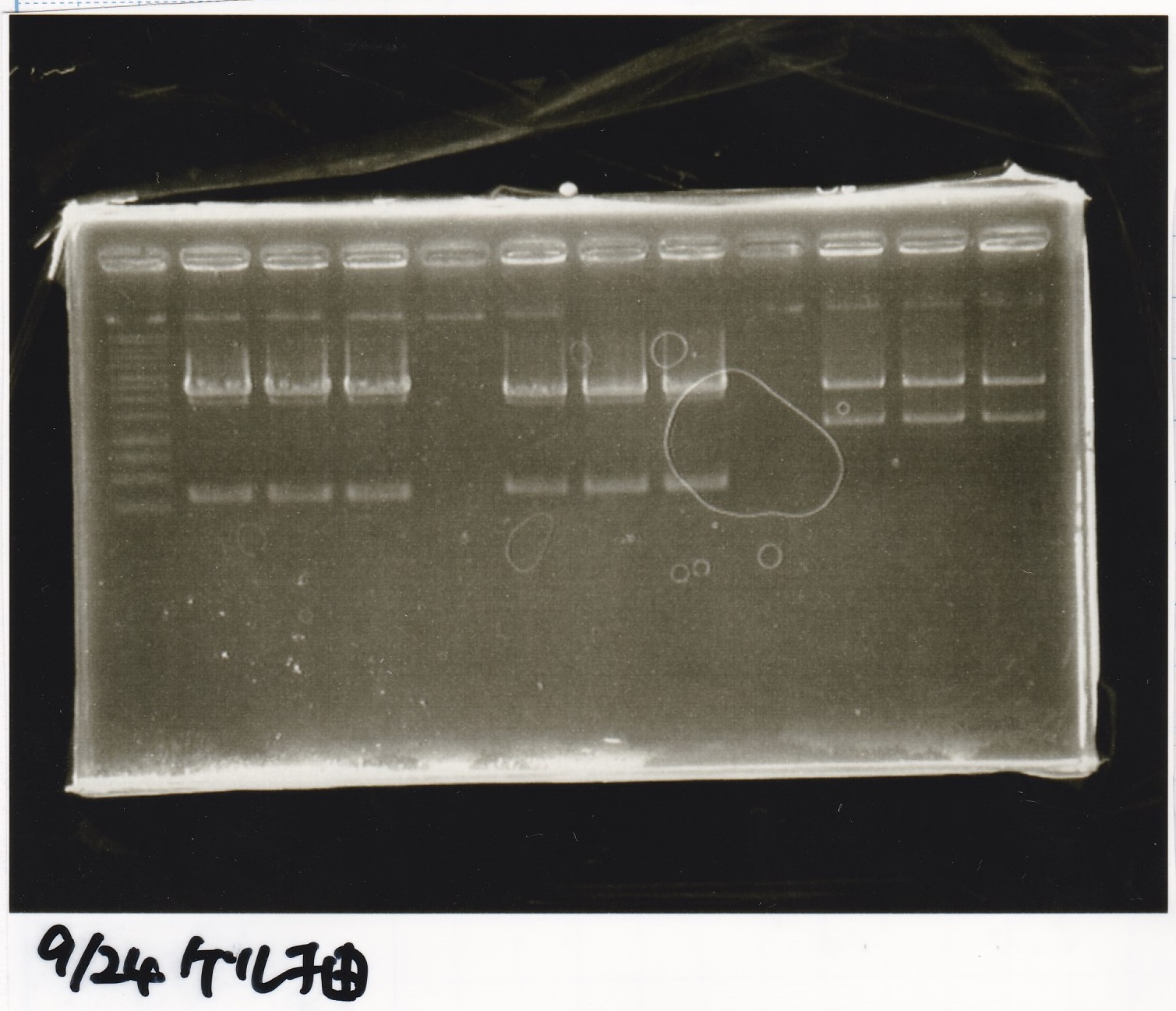
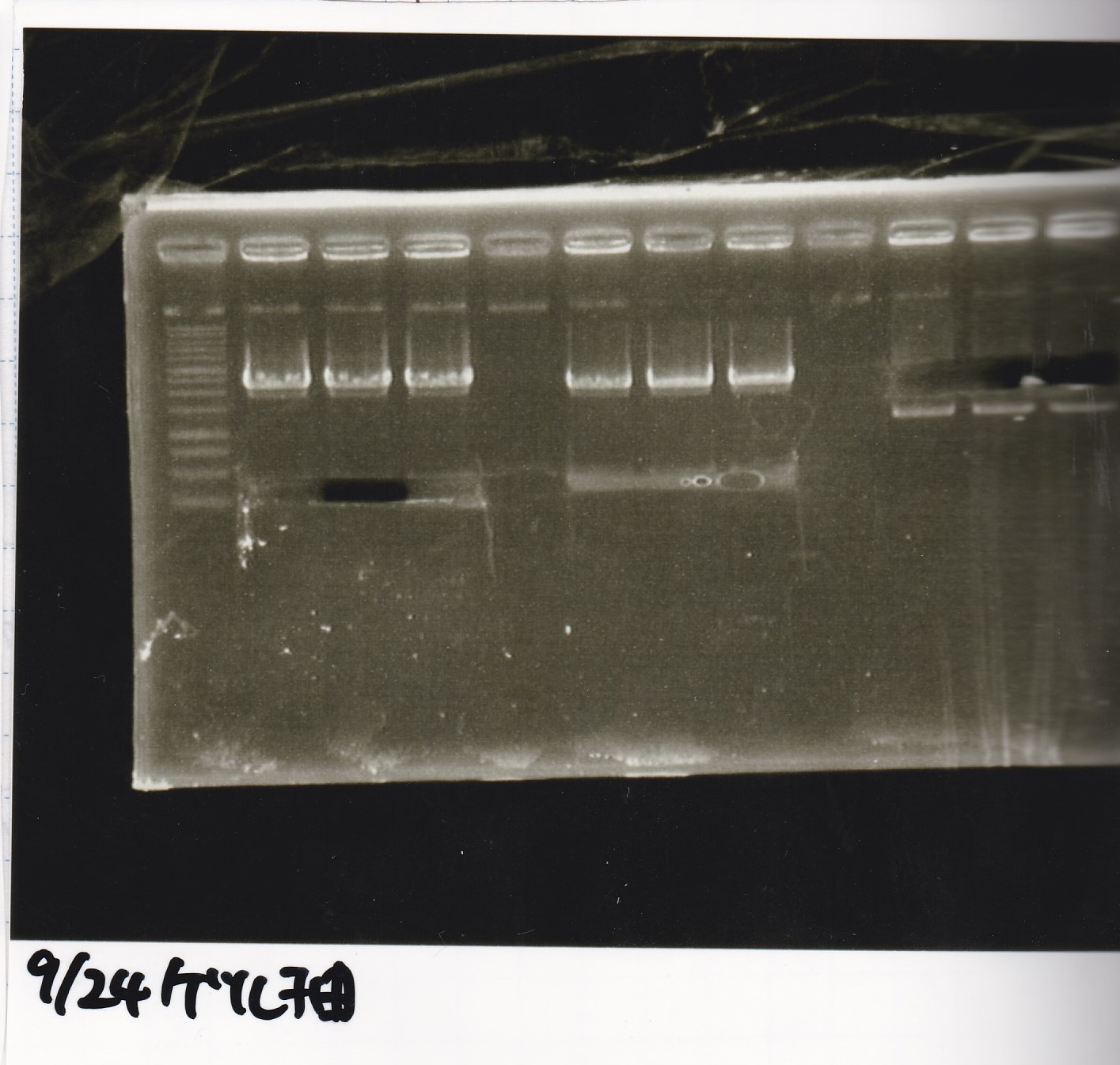
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| pT181attenuator(XbaI&PstI) | 10.1 | 2.02 | 0.43
|
| Pcon-pT181attenuator(EcoRI&SpeI) | 10.3 | 1.81 | 0.32
|
| pSB1C3(EcoRI&SpeI) | 10.0 | 2.81 | 0.02
|
Colony PCR
Nakamoto
| Sample | base pair
|
| 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 1 | 1414
|
| 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 2 | 1414
|
| 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 3 | 1414
|
| 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 4 | 1414
|
| 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 5 | 1414
|
| 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 6 | 1414
|
| 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 7 | 1414
|
| 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 8 | 1414
|
| 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 1 | 1631
|
| 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 2 | 1631
|
| 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 3 | 1631
|
| 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 4 | 1631
|
| 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 5 | 1631
|
| 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 6 | 1631
|
| 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 7 | 1631
|
| 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 8 | 1631
|
| 9/23 Pcon-RBS-tetR-DT+Pcon-RBS-GFP-DT 1 | 2042
|
| 9/23 Pcon-RBS-tetR-DT+Pcon-RBS-GFP-DT 2 | 2042
|
| 9/23 Pcon-RBS-tetR-DT+Pcon-RBS-GFP-DT 3 | 2042
|
| 9/23 Pcon-RBS-tetR-DT+Pcon-RBS-GFP-DT 4 | 2042
|
| 9/23 Pcon-pT181antisense-DT 1 | 508
|
| 9/23 Pcon-pT181antisense-DT 2 | 508
|
| 9/23 Pcon-pT181antisense-DT 3 | 508
|
| 9/23 Pcon-pT181antisense-DT 4 | 508
|
| 9/23 Pcon-pT181antisense-DT 5 | 508
|
| 9/23 Pcon-pT181antisense-DT 6 | 508
|
| 9/23 Pcon-pT181antisense-DT 7 | 508
|
| 9/23 Pcon-pT181antisense-DT 8 | 508
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 94 °C | 55 °C | 68 °C | --
|
| 5 min | 30 sec | 30 sec | 2min | 30 cycles
|
| Lane | Sample
|
| 1 | 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 1
|
| 2 | 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 2
|
| 3 | 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 3
|
| 4 | 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 4
|
| 5 | 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 5
|
| 6 | 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 6
|
| 7 | 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 7
|
| 8 | 9/23 Pcon-aptamer12-1R-DT+Pcon-RBS-GFP-DT 8
|
| 9 | 1kb ladder
|
| 10 | 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 1
|
| 11 | 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 2
|
| 12 | 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 3
|
| 13 | 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 4
|
| 14 | 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 5
|
| 15 | 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 6
|
| 16 | 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 7
|
| 17 | 9/23 Pcon-pT181attenuator-DT+Pcon-RBS-GFP-DT 8
|
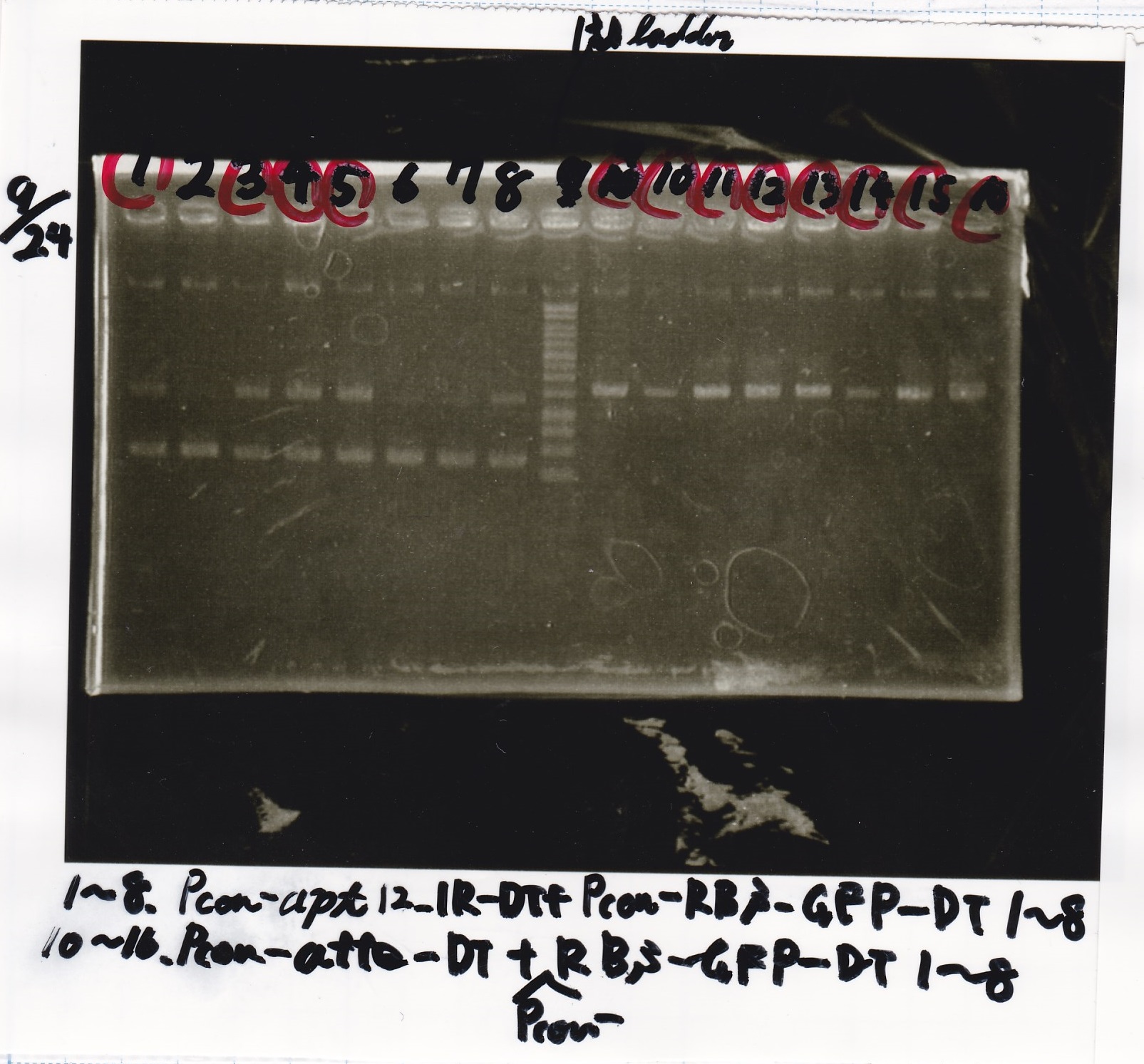
| Lane | Sample
|
| 1 | 9/23 Pcon-RBS-tetR-DT+Pcon-RBS-GFP-DT 1
|
| 2 | 9/23 Pcon-RBS-tetR-DT+Pcon-RBS-GFP-DT 2
|
| 3 | 9/23 Pcon-RBS-tetR-DT+Pcon-RBS-GFP-DT 3
|
| 4 | 9/23 Pcon-RBS-tetR-DT+Pcon-RBS-GFP-DT 4
|
| 5 | 9/23 Pcon-pT181antisense-DT 1
|
| 6 | 9/23 Pcon-pT181antisense-DT 2
|
| 7 | 1kb ladder
|
| 8 | 9/23 Pcon-pT181antisense-DT 3
|
| 9 | 9/23 Pcon-pT181antisense-DT 4
|
| 10 | 9/23 Pcon-pT181antisense-DT 5
|
| 11 | 9/23 Pcon-pT181antisense-DT 6
|
| 12 | 9/23 Pcon-pT181antisense-DT 7
|
| 13 | 9/23 Pcon-pT181antisense-DT 8
|
| 14 | --
|
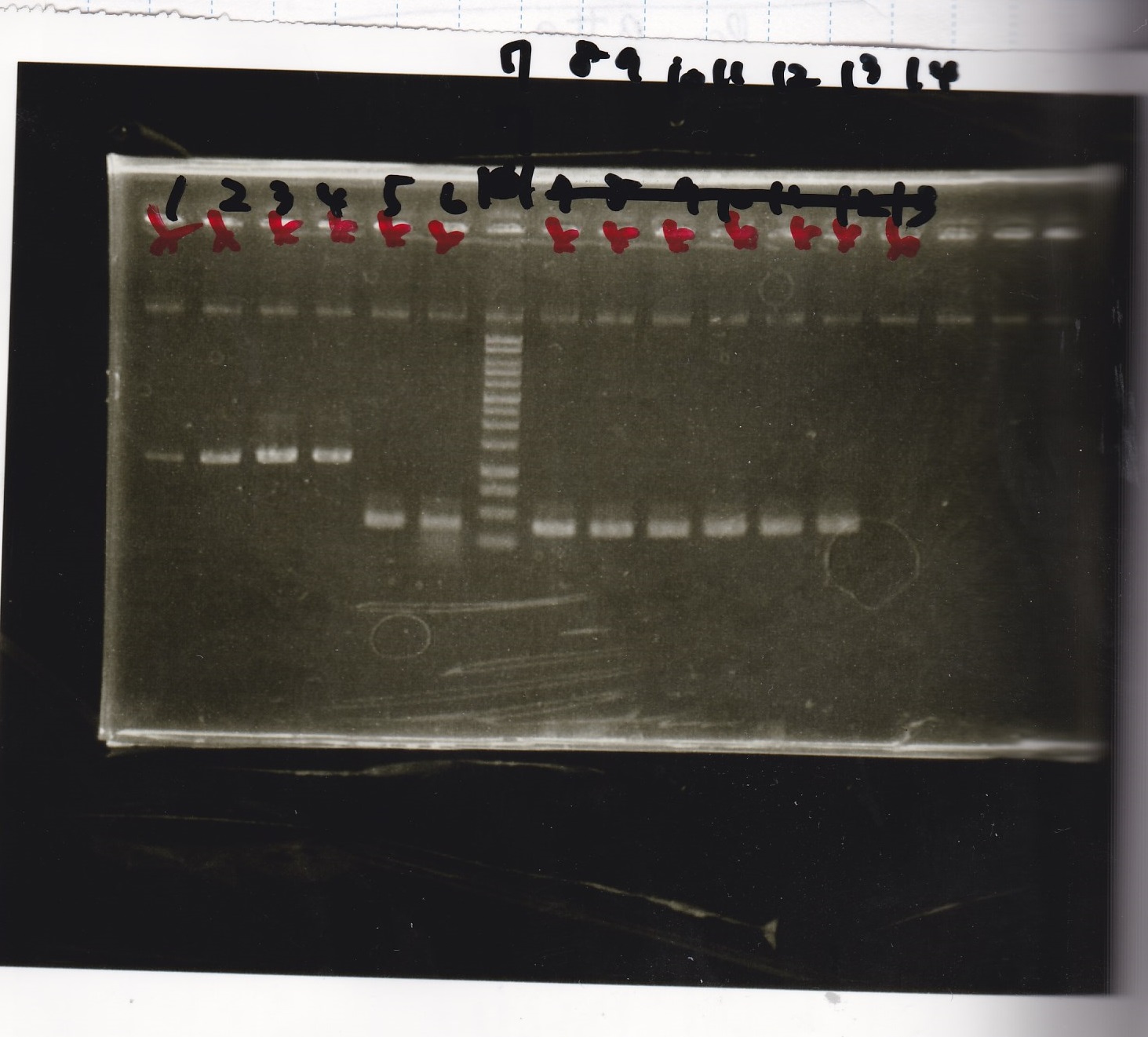
Restriction Enzyme Digestion
Nakamoto
| 9/24 aptamer12-1R-DT | EcoRI | XbaI | buffer | BSA | MilliQ | total
|
| 2cuts | 17.5µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 4.5µL | 30µL
|
| NC | 0.9µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.1µL | 10µL
|
| 9/20 Ptet | SpeI | PstI | buffer | BSA | MilliQ | total
|
| 2cuts | 9.5µL | 1.0µL | 1.0µL | 3.0µL | 0µL | 15.5µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1.0µL | 0µL | 8.5µL | 10µL
|
| 9/22 RBS-GFP-DT | EcoRI | SpeI | buffer | BSA | MilliQ | total
|
| 2cuts | 3.8µL | 1.0µL | 1.0µL | 3.0µL | 0µL | 21.2µL | 30µL
|
| NC | 0.2µL | 0µL | 0µL | 1.0µL | 0µL | 8.8µL | 10µL
|
| 9/22 Pcon-RBS-GFP-DT | EcoRI | XbaI | buffer | BSA | MilliQ | total
|
| 2cuts | 2.4µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 19.6µL | 30µL
|
| NC | 0.1µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.9µL | 10µL
|
| 9/22 Pcon-pT181attenuator 1 | EcoRI | XbaI | buffer | BSA | MilliQ | total
|
| 2cuts | 5.4µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 16.6µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.7µL | 10µL
|
| 9/22 RBS-GFP-DT | XbaI | PstI | buffer | BSA | MilliQ | total
|
| 2cuts | 3.8µL | 1.0µL | 1.0µL | 3.0µL | 3.0µL | 18.2µL | 30µL
|
| NC | 0.2µL | 0µL | 0µL | 1.0µL | 1.0µL | 7.8µL | 10µL
|
Ligation
Nakamoto
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/22 aptamer12-1R-DT(EcoRI&XbaI) | 1.4 µL | 9/24 Pcon-pT181attenuator(EcoRI&SpeI) | 26 µL | 3.5 µL
|
| experiment | 9/4 Ptet(SpeI&PstI) | 2.0 µL | 9/21 RBS-GFP-DT(XbaI&PstI) | 9.0 µL | 3.5 µL
|
- incubated at 37 °C for 1 hour.
colony PCR
Kojima
| Sample | base pair
|
| 9/23 Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 5 | 2042bp
|
| 9/23 Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 6 | 2042bp
|
| 9/23 Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 7 | 2042bp
|
| 9/23 Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 8 | 2042bp
|
| 9/23 Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 9 | 2042bp
|
| 9/23 Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 10 | 2042bp
|
| 9/23 Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 11 | 2042bp
|
| 9/23 Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 12 | 2042bp
|
| 9/23 Pcon-pT181antisense-DT 9 | 508bp
|
| 9/23 Pcon-pT181antisense-DT 10 | 508bp
|
| 9/23 Pcon-pT181antisense-DT 11 | 508bp
|
| 9/23 Pcon-pT181antisense-DT 12 | 508bp
|
| 9/23 Pcon-pT181antisense-DT 13 | 508bp
|
| 9/24 Pcon-pT181attenuator-RBS-GFP-DT 1 | 1527bp
|
| 9/24 Pcon-pT181attenuator-RBS-GFP-DT 2 | 1527bp
|
| 9/24 Pcon-pT181attenuator-RBS-GFP-DT 3 | 1527bp
|
| 9/24 Pcon-pT181attenuator-RBS-GFP-DT 4 | 1527bp
|
| 9/24 Pcon-pT181attenuator-aptamer12-1R-DT 1 | 859bp
|
| 9/24 Pcon-pT181attenuator-aptamer12-1R-DT 2 | 859bp
|
| 9/24 Pcon-pT181attenuator-aptamer12-1R-DT 3 | 859bp
|
| 9/24 Pcon-pT181attenuator-aptamer12-1R-DT 4 | 859bp
|
| 9/24 pT181attenuator(pSB1C3) 1 | 390bp
|
| 9/24 pT181attenuator(pSB1C3) 2 | 390bp
|
| 9/24 pT181attenuator(pSB1C3) 3 | 390bp
|
| 9/24 pT181attenuator(pSB1C3) 4 | 390bp
|
Tatsui
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 2min | 30cycle
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| pSB1C3 | 183.9 | 1.91 | 1.59
|
| Pcon-pT181attenuator-aptamer12-1R-DT | 231.7 | 1.82 | 2.35
|
| Plac-aptamer12-1R-DT | 208.9 | 1.98 | 1.79
|
Electrophoresis
Nakamoto
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kb ladder | -- | --
|
| 2 | 9/24 aptamer12-1R-DT | EcoRI | XbaI
|
| 3 | 9/24 aptamer12-1R-DT | -- | --
|
| 4 | 9/24 Ptet | SpeI | PstI
|
| 5 | 9/24 Ptet | -- | --
|
| 6 | 9/24 RBS-GFP-DT | EcoRI | SpeI
|
| 7 | 9/24 RBS-GFP-DT | -- | --
|
| 8 | 9/24 Pcon-RBS-GFP-DT | EcoRI | XbaI
|
| 8 | 9/24 Pcon-RBS-GFP-DT | -- | --
|
| 9 | 9/24 Pcon-RBS-GFP-DT | -- | --
|
| 10 | 9/24 Pcon-pT181attenuator 1 | EcoRI | XbaI
|
| 11 | 9/24 Pcon-pT181attenuator 1 | -- | --
|
| 12 | 9/24 RBS-GFP-DT | XbaI | PstI
|
| 13 | 9/24 RBS-GFP-DT | -- | --
|
| 14 | 1kb ladder | -- | --
|
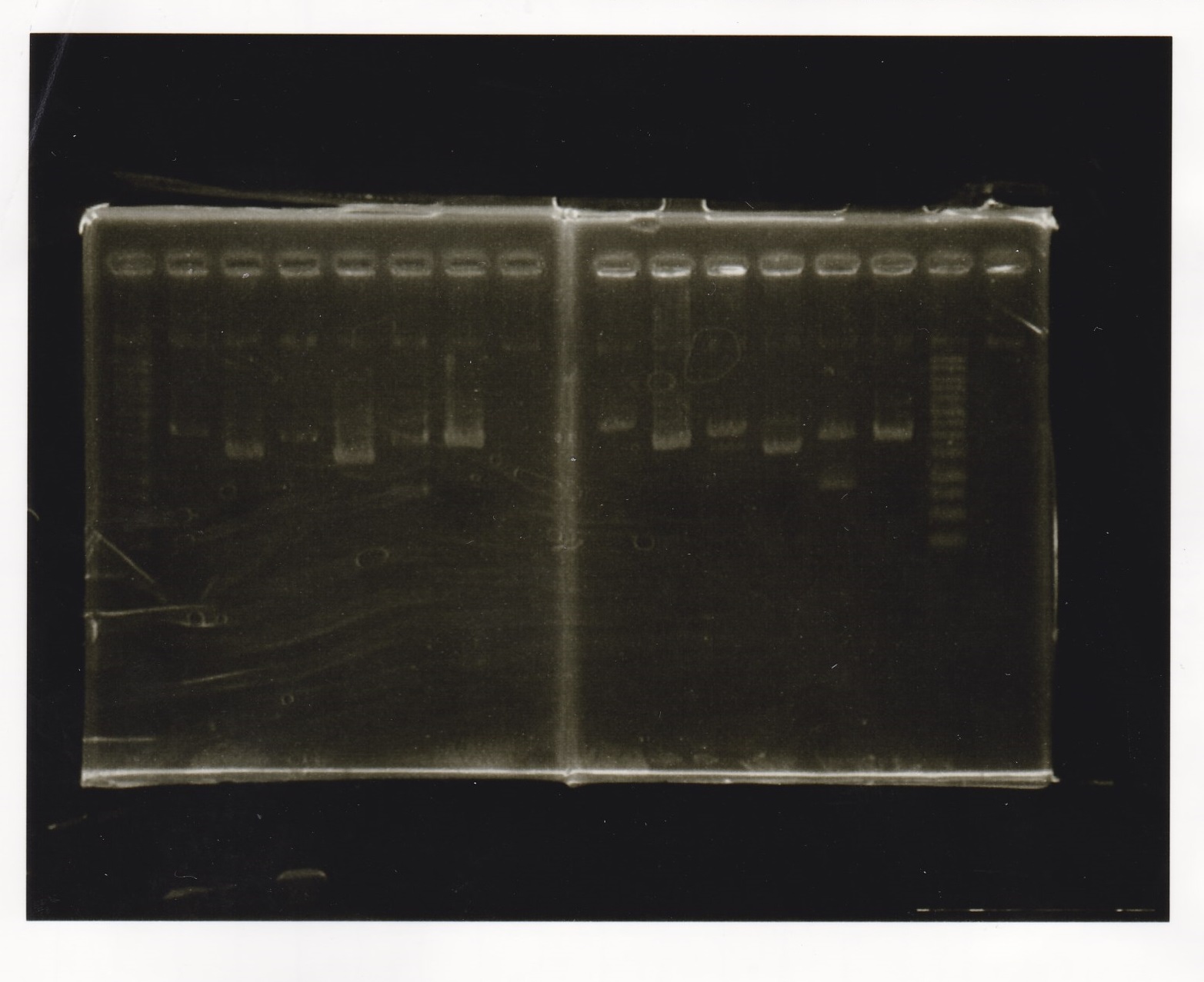
No name
1
| Lane | DNA | Enzyme1 | Enzyme2
|
| 1 | 1kb ladder | -- | --
|
| 2 | aptamer12-1R-DT | EcoRI | XbaI
|
| 3 | aptamer12-1R-DT | EcoRI | XbaI
|
| 4 | aptamer12-1R-DT | EcoRI | XbaI
|
| 5 | -- | -- | --
|
| 6 | Ptet | SpeI | PstI
|
| 7 | Ptet | SpeI | PstI
|
| 8 | Ptet | SpeI | PstI
|
| 9 | -- | -- | --
|
| 10 | RBS-GFP-DT | EcoRI | SpeI
|
| 11 | RBS-GFP-DT | EcoRI | SpeI
|
| 12 | RBS-GFP-DT | EcoRI | SpeI
|
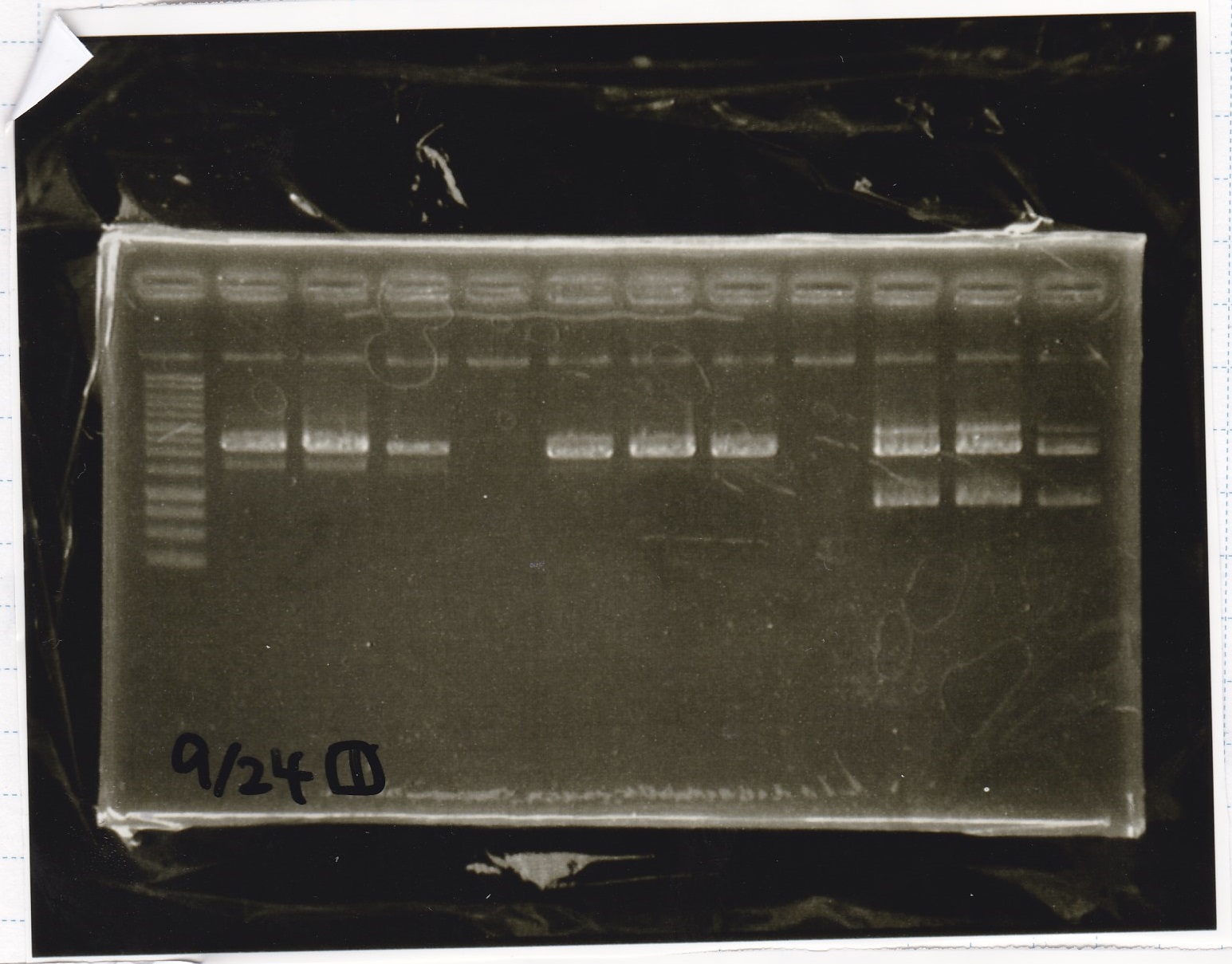

| Lane | DNA | Enzyme1 | Enzyme2
|
| 1 | 1kb ladder | -- | --
|
| 2 | Pcon-RBS-GFP-DT | EcoRI | XbaI
|
| 3 | Pcon-RBS-GFP-DT | EcoRI | XbaI
|
| 4 | Pcon-RBS-GFP-DT | EcoRI | XbaI
|
| 5 | -- | -- | --
|
| 6 | Pcon-pT181attenuator | EcoRI | XbaI
|
| 7 | Pcon-pT181attenuator | EcoRI | XbaI
|
| 8 | Pcon-pT181attenuator | EcoRI | XbaI
|
| 9 | -- | -- | --
|
| 10 | RBS-GFP-DT | XbaI | PstI
|
| 11 | RBS-GFP-DT | XbaI | PstI
|
| 12 | RBS-GFP-DT | XbaI | PstI
|
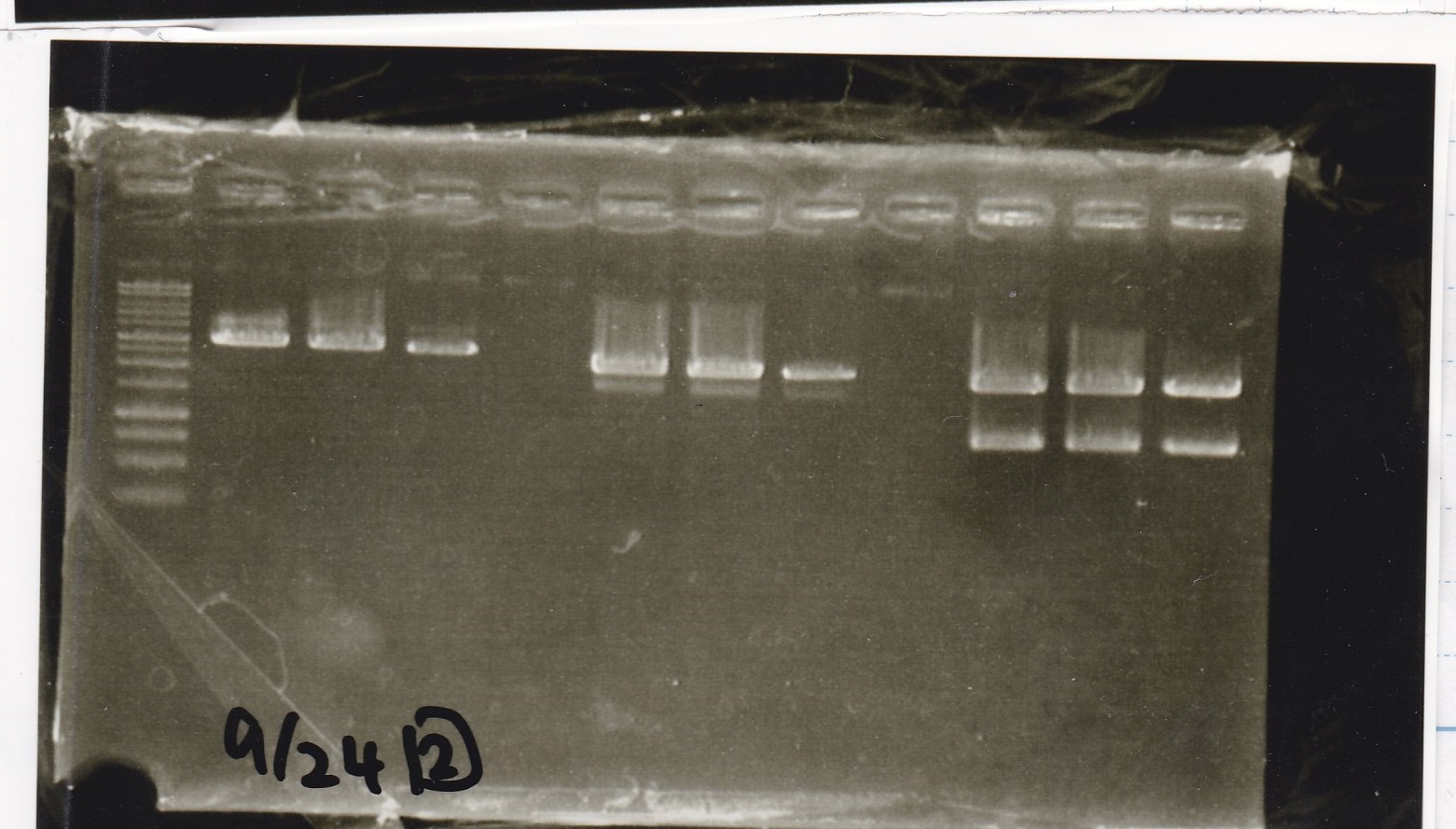

Electrophoresis
No name
1
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 5
|
| 3 | Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 6
|
| 4 | Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 7
|
| 5 | Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 8
|
| 6 | Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 9
|
| 7 | Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 10
|
| 8 | 1kb ladder
|
2
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 11
|
| 3 | Pcon-RBS-tetR-DT-Pcon-RBS-GFP-DT 12
|
| 4 | Pcon-pT181attenuator-RBS-GFP-DT 1
|
| 5 | Pcon-pT181attenuator-RBS-GFP-DT 2
|
| 6 | Pcon-pT181attenuator-RBS-GFP-DT 3
|
| 7 | Pcon-pT181attenuator-RBS-GFP-DT 4
|
| 8 | 1kb ladder
|
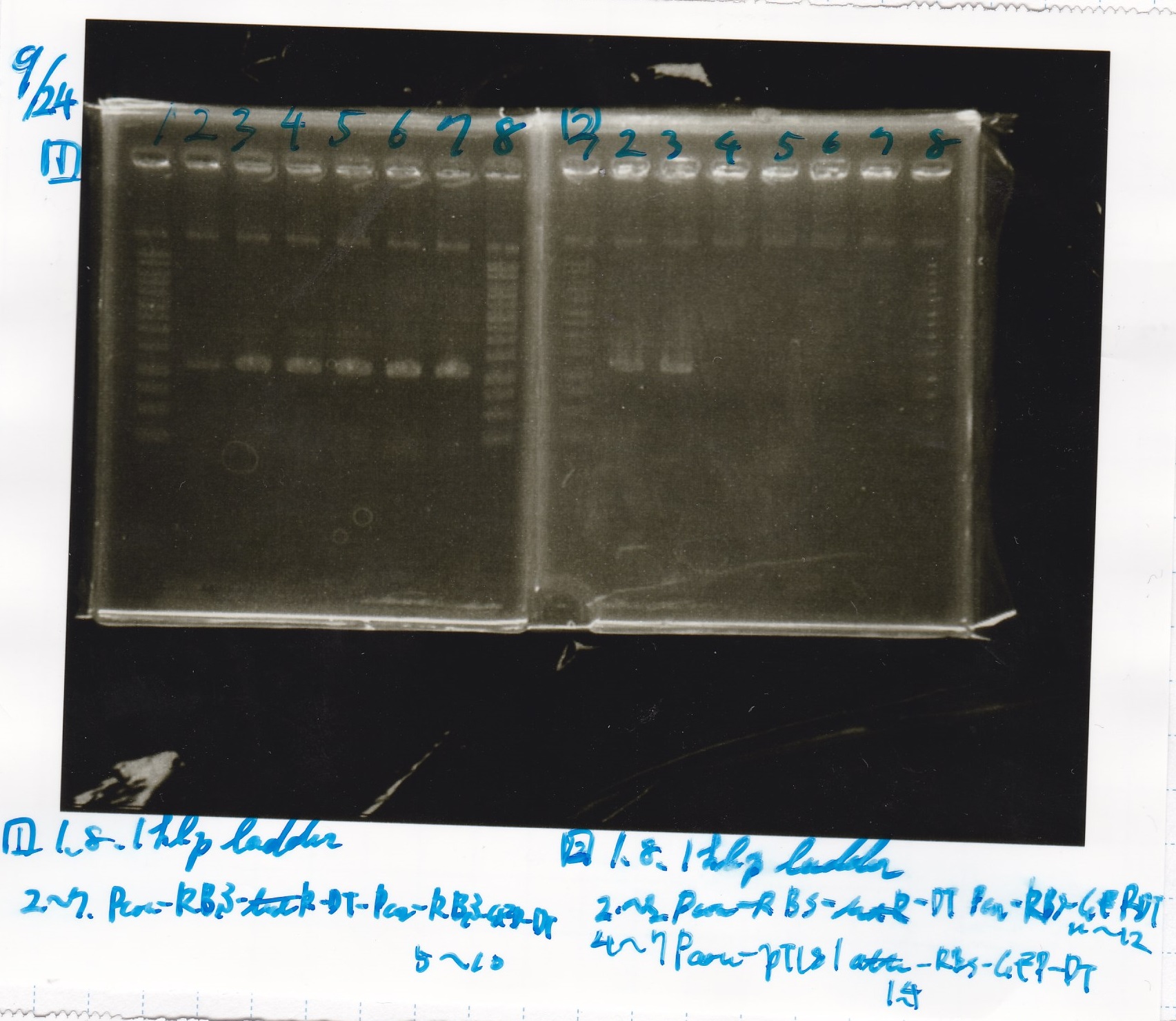
3
| Lane | Sample
|
| 1 | Pcon-pT181antisense-DT 9
|
| 2 | Pcon-pT181antisense-DT 10
|
| 3 | Pcon-pT181antisense-DT 11
|
| 4 | Pcon-pT181antisense-DT 12
|
| 5 | Pcon-pT181antisense-DT 13
|
| 6 | 100bp ladder
|
| 7 | Pcon-pT181attenuator-aptamer12-1R-DT 1
|
| 8 | Pcon-pT181attenuator-aptamer12-1R-DT 2
|
4
| Lane | Sample
|
| 1 | Pcon-pT181attenuator-aptamer12-1R-DT 3
|
| 2 | Pcon-pT181attenuator-aptamer12-1R-DT 4
|
| 3 | 100bp ladder
|
| 4 | pT181attenuator(pSB1C3) 1
|
| 5 | pT181attenuator(pSB1C3) 2
|
| 6 | pT181attenuator(pSB1C3) 3
|
| 7 | pT181attenuator(pSB1C3) 4
|
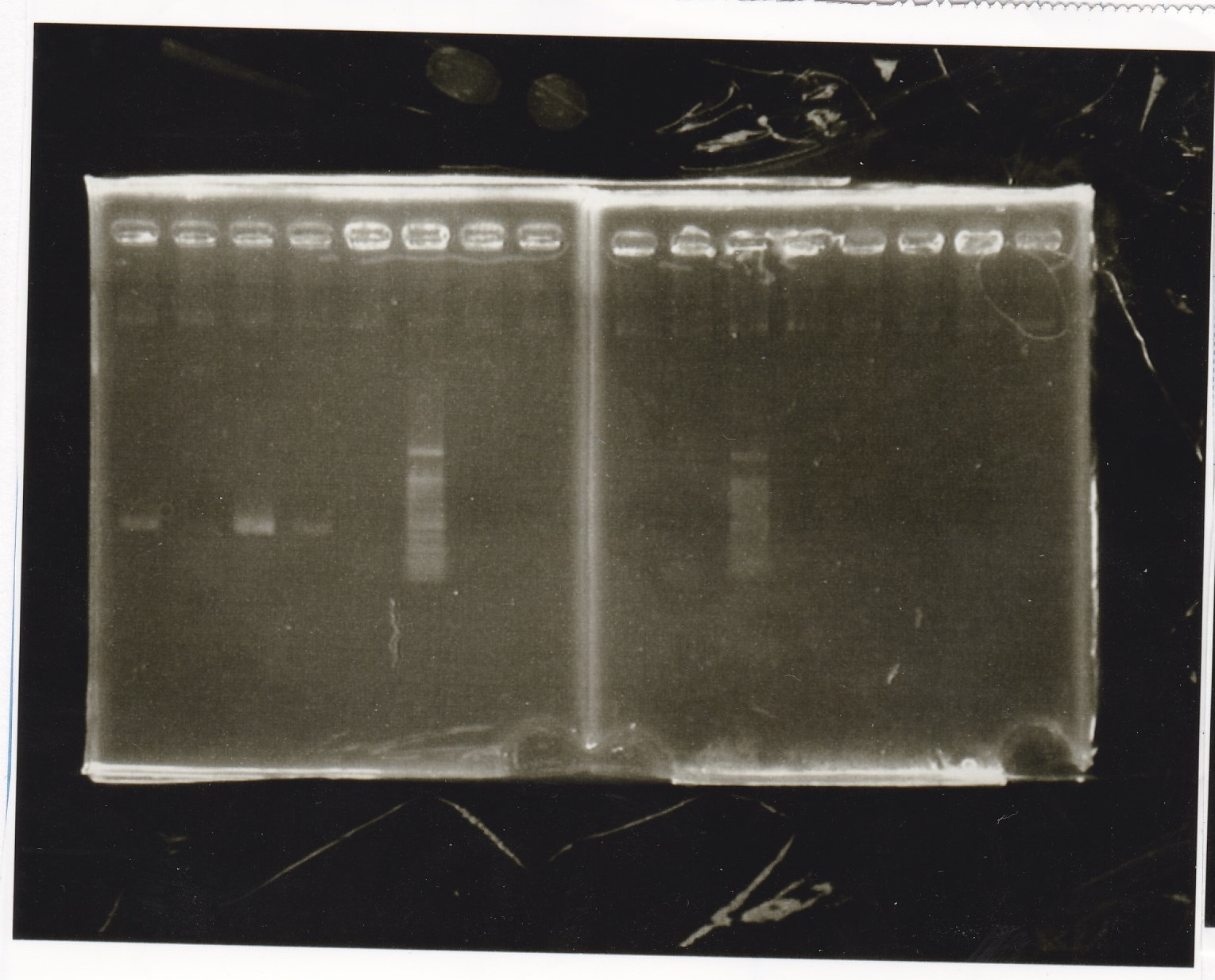
Ligation
Hirano
| state | Vector | Inserter1 | Inserter2 | Inserter3 | Inserter4 | (NEB)T4ligase | (NEB)Buffer | milliQ | total
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | tetR-aptamer12-1R-DT(1-3) | 1 µL | Ptet(2-S) | 0.3µL | Pcon-tetR-aptamer12-1R-DT(3-2A) | 1.1µL | Plac(E-1A) | 0.5µL | 0.5µL | 0.5µL | 0.2 µL | 5µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-pT181attenuator(E-1A) | 1 µL | Pcon-tetR-aptamer12-1R-DT(1-2A) | 1µL | Ptet(2-S) | 0.3µL | -- | -- | 0.5µL | 0.5µL | 0.7 µL | 5µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-tetRaptamer12-1R-DT(E-1A) | 2µL | Pcon-tetRaptamer12-1R-DT(1-2A) | 1µL | Ptet(2-S) | 0.3µL | -- | -- | 0.5µL | 0.5µL | 0 µL | 5.3µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-spinach-DT(E-1A) | 3µL | Pcon-tetRaptamer12-1R-DT(1-2A) | 1µL | Ptet(2-S) | 0.3µL | -- | -- | 0.5µL | 0.5µL | 0 µL | 6.3µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-pT181antisense-DT(E-1A) | 0.5µL | DT(1-3) | 2µL | Ptet(2-S) | 0.3µL | Pcon-tetRaptamer12-1R-DT(3-2A) | 1.1µL | 0.5µL | 0.5µL | 0.1 µL | 5µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-pT181attenuator(E-1A) | 1µL | tetRaptamer12-1R-DT(1-3) | 1µL | Ptet(2-S) | 0.3µL | Pcon-tetRaptamer12-1R-DT(3-2A) | 1.1µL | 0.5µL | 0.5µL | 0µL | 5.4µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Ptet(2-S) | 0.3µL | Pcon-tetRaptamer12-1R-DT(E-2A) | 1µL | -- | -- | -- | -- | 0.5µL | 0.5µL | 1.7µL | 5µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-GFP-DT(2-SA) | 0.5µL | 1µL | -- | -- | -- | -- | -- | 0.5µL | 0.5µL | 1.5µL | 5µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-tetRaptamer12-1R-DT(E-1A) | 2µL | Pcon-pT181attenuator(1-SA) | 2.1µL | -- | -- | -- | -- | 0.5µL | 0.5µL | 0µL | 6.1µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-pT181attenuator-DT(E-1A) | 1µL | Pcon-pT181attenuator(1-SA) | 2.1µL | -- | -- | -- | -- | 0.5µL | 0.5µL | 0µL | 5.1µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-tetR12-1R-DT(E-1A) | 2µL | Pcon-GFP-DT(2-SA) | 0.5µL | spinach-DT(3-2) | 1µL | Ptet-pT181antisense(1-3) | 1µL | 0.5µL | 0.5µL | 0µL | 6.5µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-tetRaptamer12-1R-DT(E-1A) | 2µL | Pcon-pT181antisense(1-2A) | 0.7µL | Pcon-pT181attenuator(2-SA) | 2.1µL | -- | -- | 0.5µL | 0.5µL | 0µL | 6.8µL
|
| experiment | RBS-GFP-DT(pSB1C3) | 1µL | Pcon-tetRaptamer12-1R-DT(E-1A) | 2µL | Pcon-pT181attenuator(2-SA) | 2.1µL | Spinach-DT(3-2) | 1µL | Ptet-pT181antisense(1-3) | 1µL | 0.5µL | 0.5µL | 0µL | 8.1µL
|
Concentrate total (more than 5µL) by evaporater until volume become less than 5 µL
PCR
No name
| genome DNA | KOD plus | 10x buffer | dNTP | MgSO4 | F primer | R primer | KOD-plus | MilliQ | total
|
| Pcon antisense(1C3-GGE-fwd,1A2-GG2-rev) | 1µL | 2.5µL | 2.5µL | 1.5µL | 0.75µL | 0.75µL | 0.5µL | 15.5 | 25µL
|
| DT-(1C3-GG1-fwd,1C3-GG2-rev) | 1µL | 2.5µL | 2.5µL | 1.5µL | 0.75µL | 0.75µL | 0.5µL | 15.5 | 25µL
|
| Spinach-DT-(1C3-GG1-fwd,1C3-GG2-rev) | 1µL | 2.5µL | 2.5µL | 1.5µL | 0.75µL | 0.75µL | 0.5µL | 15.5 | 25µL
|
| Pcon-antisense-spinach-DT(1C3-GGE-fwd,1A2-GG1-rev) | 1µL | 2.5µL | 2.5µL | 1.5µL | 0.75µL | 0.75µL | 0.5µL | 15.5 | 25µL
|
| Ptet(1C3-GGE-fwd,1C3-GG0-rev) | 1µL | 2.5µL | 2.5µL | 1.5µL | 0.75µL | 0.75µL | 0.5µL | 15.5 | 25µL
|
| Pcon attenuator(1C3-GGE-fwd,1C3-GG0-rev) | 1µL | 2.5µL | 2.5µL | 1.5µL | 0.75µL | 0.75µL | 0.5µL | 15.5 | 25µL
|
| DT(1C3-GG2-fwd,1C3-GG1-rev) | 1µL | 2.5µL | 2.5µL | 1.5µL | 0.75µL | 0.75µL | 0.5µL | 15.5 | 25µL
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10sec | 30sec | 36sec | 30
|
Sep 25
Electrophoresis
nakamoto
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Pcon-antisense (1C3-66E-fwd A2-662 rev) | -- | --
|
| 3 | DT (1C3-661-fwd 1C3 662 rev) | -- | --
|
| 4 | spinach-DT(1C3-661-fwd 1C3 662 rev) | -- | --
|
| 5 | Pcon-antisense-spinach-DT(1C3-66E fwd A2-661 rev) | -- | --
|
| 6 | 100bp ladder | -- | --
|
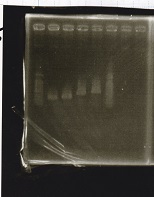
| Lane | Sample | Enzyme1 | Enzyme2
|
| 2 | 100bp ladder | -- | --
|
| 3 | Ptet (1C3 66E fwd-1C3 660 rev) | -- | --
|
| 4 | Pcon-attenuator(1C3-660 fwd A2-661 rev) | -- | --
|
| 5 | DT(1C3 662 fwd-1C3 661 rev) | -- | --
|
| 6 | Pcon-attenuator-aptamer-DT(1C3 66E fwd-A2-661-rev) | -- | --
|
| 7 | 100bp ladder | -- | --
|

Colony PCR
Nakamoto
| Sample | base pair
|
| 9/23 Pcon-PT181 attenuator-aptamer12-1R-DT-(5~12) | 859
|
| 9/23 Pcon-PT181 attenuator-RBS-GFP-DT-(5~8) | 1527
|
| 9/23 Pcon-PT181 attenuator-pSB1C3-(5~8) | 601
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94 °C | 94 °C | 55 °C | 68 °C | --
|
| 2 min | 30 s | 30 s | 1min5s | 30 cycles
|
Electrophoresis
no name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | Pcon-PT181 attenuator-aptamer12-1R-DT-5 | -- | --
|
| 2 | Pcon-PT181 attenuator-aptamer12-1R-DT-6 | -- | --
|
| 3 | Pcon-PT181 attenuator-aptamer12-1R-DT-7 | -- | --
|
| 4 | Pcon-PT181 attenuator-aptamer12-1R-DT-8 | -- | --
|
| 5 | Pcon-PT181 attenuator-aptamer12-1R-DT-9 | -- | --
|
| 6 | Pcon-PT181 attenuator-aptamer12-1R-DT-10 | -- | --
|
| 7 | Pcon-PT181 attenuator-aptamer12-1R-DT-11 | -- | --
|
| 8 | Pcon-PT181 attenuator-aptamer12-1R-DT-12 | -- | --
|
| 9 | 1kbp ladder | -- | --
|
| 10 | Pcon-PT181 attenuator-RBS-GFP-DT-5 | -- | --
|
| 11 | Pcon-PT181 attenuator-RBS-GFP-DT-6 | -- | --
|
| 12 | Pcon-PT181 attenuator-RBS-GFP-DT-7 | -- | --
|
| 13 | Pcon-PT181 attenuator-RBS-GFP-DT-8 | -- | --
|
| 14 | Pcon-PT181 attenuator-pSB1C3-5 | -- | --
|
| 15 | Pcon-PT181 attenuator-pSB1C3-6 | -- | --
|
| 16 | Pcon-PT181 attenuator-pSB1C3-7 | -- | --
|
| 17 | Pcon-PT181 attenuator-pSB1C3-8 | -- | --
|
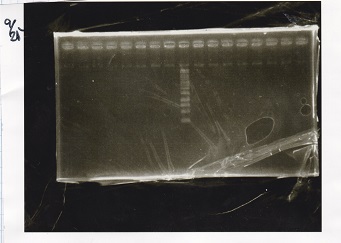
Transformation
kojima and nakamoto
| Name | Sample | Competent Cells | Total | Plate
|
| Plac+tet aptamer12+Pcon tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-pT181attenuator-DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-tet aptamer12-DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-Spinach-DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-pT181antisense+DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-pT181attenuator+tet aptamer12-DT+Pcon-tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon tetR-DT+Ptet+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon tetR-DT+Pcon-GFP-DT+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-tet aptamer12-DT+Pcon-pT181attenuator+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-pT181attenuator-DT+Pcon-pT181attenuator+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-tet aptamer12-DT+Ptet-pT181antisense+Spinach-DT+Pcon-RBS-GFP-DT+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-tet aptamer12-DT+Pcon-pT181antisense+Spinach-DT+Pcon-pT181attenuator+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Pcon-tet aptamer12-DT+Ptet-pT181antisense+Spinach-DT+Pcon-pT181attenuator+RBS-GFP-DT | 1µL | 10µL | 11µL | --
|
| Name | Sample | Competent Cells | Total | Plate
|
| aptamer12-DT(EcoRI+XbaI)+Pcon-pT181attenuator(EcoRI+SpeI) | 2µL | 20µL | 22µL | --
|
| Ptet(SpeI+PstI)+RBS-GFP-DT(XbaI+PstI)) | 2µL | 20µL | 22µL | --
|
Ligation
nakamoto
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/25 aptamer12_1R-DT(EcoRI&XbaI) | 4.5 µL | Pcon-pT181 attenuator(EcoRI&SpeI) | 7.7 µL | 6.1 µL
|
| experiment | 9/17 pSB1C3(EcoRI&SpeI) | 2.0 µL | 9/3 pT181 attenuator(EcoRI&SpeI) | 14 µL | 8 µL
|
| experiment | 9/25 Pcon-pT181 attenuator(EcoRI&XbaI) | 4.5 µL | 9/25 RBS-GFP-DT(EcoRI&SpeI) | 19.3 µL | 11.9 µL
|
| experiment | 9/25 Pcon-RBS-GFP-DT(EcoRI&XbaI) | 4.2 µL | Pcon-RBS-tetR-DT(EcoRI&SpeI) | 20 µL | 12.1 µL
|
Restriction Enzyme Digestion
nakamoto>
| | 9/24 Pcon-attenuator-aptamer-DT(µL) | EcoRI(µL) | SpeI(µL) | Buffer(µL) | MilliQ(µL) | total(µL)
|
| 2 cuts | 8.6 | 1 | 1 | 3 | 16.4 | 30
|
| NC | 0.4 | 0 | 0 | 1 | 8.6 | 10
|
| | 9/11 DT(µL) | EcoRI(µL) | XbaI(µL) | Buffer(µL) | BSA(µL) | MilliQ(µL) | total(µL)
|
| 2 cuts | 10.4 | 1 | 1 | 3 | 3 | 11.6 | 30
|
| NC | 0.5 | 0 | 0 | 1 | 1 | 7.5 | 10
|
Liquid Culture
Kojima
| Sample | medium
|
| DT | --
|
| pT181attenuator | --
|
| Pcon-RBS-tetR-DT | --
|
Kojima
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 2 | Pcon-pT181antisense | --
|
| 3
|
| 4
|
| 5 | -- | --
|
| 6 | DT | --
|
| 7
|
| 8
|
| 9 | -- | --
|
| 10 | Spinach-DT | --
|
| 11
|
| 12
|
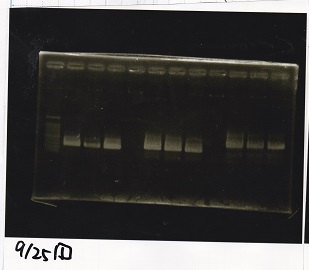
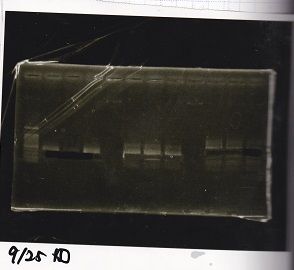
| Lane | DNA | Enzyme
|
| 1 | 100bp ladder | --
|
| 3 | Pcon-antisense-spinach-DT(E-1A) | --
|
| 4 | Pcon-antisense-spinach-DT(E-1A) | --
|
| 5 | Pcon-antisense-spinach-DT(E-1A) | --
|
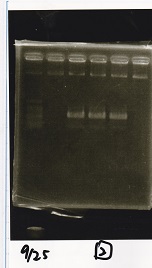
| Lane | DNA | Enzyme
|
| 7 | 100bp ladder | --
|
| 9 | Ptet(E-0) | --
|
| 10 | Ptet(E-0) | --
|
| 11 | Ptet(E-0) | --
|

| Lane | DNA | Enzyme
|
| 13 | 100bp ladder | --
|
| 15 | Pcon-attenuator(0-1A) | --
|
| 16 | Pcon-attenuator(0-1A) | --
|
| 17 | Pcon-attenuator(0-1A) | --
|
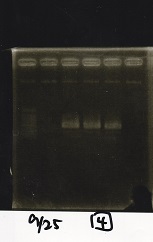
||--
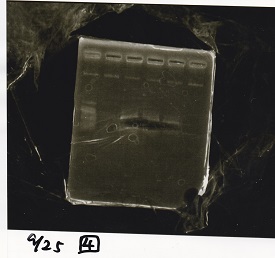
| Lane | DNA | Enzyme
|
| 19 | 100bp ladder | --
|
| 21 | Pcon-attenuator-aptamer-DT(E-1A) | --
|
| 22 | Pcon-attenuator-aptamer-DT(E-1A) | --
|
| 23 | Pcon-attenuator-aptamer-DT(E-1A) | --
|
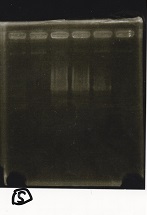
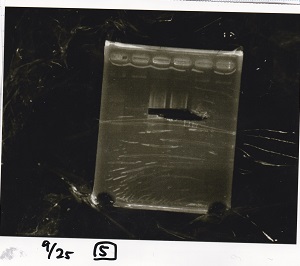
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| DT(2-1) | 24.9 | 1.73 | 0.37
|
| Ptet(E-0) | 12.8 | 1.59 | 0.90
|
| Pcon-antisense(E-2A) | 16.5 | 1.72 | 1.07
|
| Pcon-attenuator-aptamer-DT(E-1A) | 22.1 | 1.75 | 0.94
|
| Pcon-antisense-spinach-DT(E-1A) | 31.0 | 1.78 | 1.15
|
| spinach-DT(1-2) | 23.4 | 1.85 | 1.11
|
| Pcon-attenuator(0-1A) | 22.5 | 1.76 | 0.46
|
| DT(1-2) | 25.1 | 1.78 | 1.21
|
Electrophoresis
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kbp ladder | -- | --
|
| 2 | Pcon-pT181 attenuator-aptamer 12_1R-DT | EcoRI | SpeI
|
| 3 | Pcon-pT181 attenuator-aptamer 12_1R-DT | -- | --
|
| 4 | DT | EcoRI | XbaI
|
| 5 | DT | -- | --
|
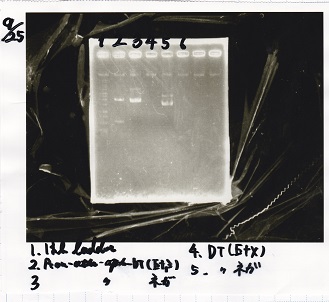
Nakamoto
| Lane | DNA | Enzyme
|
| 1 | DT | EcoRI+XbaI
|
| 2 | DT | EcoRI+XbaI
|
| 3 | Pcon-pT181 attenuator-aptamer12_1R-DT | EcoRI+SpeI
|
| 4 | Pcon-pT181 attenuator-aptamer12_1R-DT | EcoRI+SpeI
|
| 5 | Pcon-pT181 attenuator-aptamer12_1R-DT | EcoRI+SpeI
|

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-pT181 attenuator-aptamer12_1R-DT(EcoRI&SpeI) | 11.8 | 1.80 | 0.46
|
| DT(EcoRI+XbaI) | 42.6 | 1.84 | 0.98
|
Liquid Culture
No name
| Sample | medium
|
| Pcon-pT181anntisense-spinach-DT(4k5) | -
|
Ligation
Nakamoto and Tatsui
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/24 pSB1C3(EcoRI+Spel) | 10µL | 9/25 PconpT181attenuator-aptamer12-1R-DT(EcoRI+Spel) | 5.9µL | 8µL
|
PCR
Hirano
| 800pg/μL DT | KOD plus | 10x buffer | dNTP | MgSO4 | 1C3_GG2_fwd | iC3_GG1_rev | MilliQ | total
|
| 1 | 0.5 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 57°C | 68°C | --
|
| 2min | 10s | 30s | 24s | 30cycles
|
Transformation
Tatsui
| Name | Plate
|
| aptamer12-1R-DT | CP
|
| pSB1C3(EcoRI+SpeI)+pT181attenuator(EcoRI+Spal) | CP
|
| Pcon-pT181attenuator(EcoRI+XbaI)+RBS-GFP-DT(EcoRI+SpeI) | Amp
|
| Pcon-RBS-GFP-DT(EcoRI+XbaI)+Pcon-RBS-tetR-DT(EcoRI+SpeI) | Amp
|
| pSB1C3(EcoRI+SpeI)+Pcon-pT181attenuator-aptamer12-1R-DT(EcoRI+SpeI) | CP
|
Electrophoresis
Hirano
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | DT(2-1) | -- | --
|

Column Refining
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 8/25 DT(2-1) | -- | -- | --
|
Restriction Enzyme Digestion
Concentrate PCR product by evaporator until volume become less than 3µL, add all sulution and pipetting.
| 9/25 DT(2-1) | 10*cut smart Buffer | BsaI-HF | MilliQ | total
|
| 3.0 µL | 2.0 µL | 0.3 µL | 14.7 µL | 20.0 µL
|
Incubate 37°C 12hour
Liquid Culture
Hirano
| Sample | medium
|
| Pcon-GFP-DT | LB(Amp)
|
| RBS-GFP-DT | LB(CP)
|
| Pcon-spinach-DT | LB(Amp)
|
| Pcon-tetRaptamer-DT | LB(Amp)
|
| Pcon-attenuator-DT | LB(Amp)
|
| spinach-DT | LB(CP)
|
| Pcon-antisense-spinach-DT | LB(Amp)
|
Plating
Hirano
Mix Liquid Culture 1mL and LB(+Amp), incubate 37°C 30min
OD600-1.445
| Sample | Use plate | The number of plate
|
| Pcon-GFP-DT | M9(+Amp) | 3
|
| Pcon-GFP-DT | LB(+Amp) | 3
|
</div>
incubate 37°C
Colony PCR
No name
| Sample | base pair
|
| 9/24 GGA-1(1~8) | 3177
|
| 9/24 GGA-2(1~8) | 3189
|
| 9/24 GGA-3(1~8) | 2989
|
| 9/24 GGA-4(1~8) | 2939
|
| 9/24 GGA-7(1) | 2489
|
| GGA-16(1) | 2717
|
| RBS-GFP-DT | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 3min15s | 30cycles
|
Nakamoto
sumple 0.25mL
Add 1mLISOGEN-LS
Lysis or homogenization
Store for 5 min, at room temperature
Add 0.2mL Chloroform
Shake vigorously for 15sec.
Store for 2~3min.at room temperature
Centrifuge 12K*g for 15min. at 4°C
Extract aqueous phase
Add 0.5mL isopropanol
Store for 5~10min. at room temperature
Centrifuge 12K*g for 10min. at 4°C
Remove aqueous phase
Add at least 1mL 70% ethanol
Vortex
Centrifuge 7.5K*g for 5min. at 4°C
Remove aqueous phase
Dry briefly
Add ddH2O, TE(pH8.0) or 0.5% SDS
Dissolve
Total RNA solution
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-RBS-GFP-DT | 2783 | 1.95 | 1.48
|
Add 91.2µL, Store at freezer
Total 250µL</div>
cDNA Synthesis
Nakamoto
| volume | 7 µL
|
| 7*cDNA Wipcout Buffer | 1 µL
|
| Template RNA(9/25 Pcon-RBS-GFP-DT) | 3 µL
|
| RNase-frec water | 4 µL
|
Incubate 42°C 2min, then on ice immediately
Add
| 5*Quantiscript Reverse Transcriptase | 0.5 µL
|
| 5*Quantiscript RT Buffer | 2 µL
|
| RT Primer Mix | 0.5µL
|
Incubate 42°C 15min, then incubate 95°C 3min
Electrophoresis
Nakamoto
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 100bp ladder | -- | --
|
| 2 | Pcon-RBS-GFP-DT(RNA) | -- | --
|

Ligation
Tatsui
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/13 Pcon-pT181-attenuator(SpeI&PstI) | 3.5µL | 9/25 RBS-GFP-DT(XbaI&PstI) | 13.5µL | 8.5 µL
|
Sep 26
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | GGA1-1 | -- | --
|
| 2 | GGA1-2 | -- | --
|
| 3 | GGA1-3 | -- | --
|
| 4 | GGA1-4 | -- | --
|
| 5 | GGA1-5 | -- | --
|
| 6 | GGA1-6 | -- | --
|
| 7 | GGA1-7 | -- | --
|
| 8 | GGA1-8 | -- | --
|
| 9 | 1kbp ladder | -- | --
|
| 10 | GGA2-1 | -- | --
|
| 11 | GGA2-2 | -- | --
|
| 12 | GGA2-3 | -- | --
|
| 13 | GGA2-4 | -- | --
|
| 14 | GGA2-5 | -- | --
|
| 15 | GGA2-6 | -- | --
|
| 16 | GGA2-7 | -- | --
|
| 17 | GGA2-8 | -- | --
|
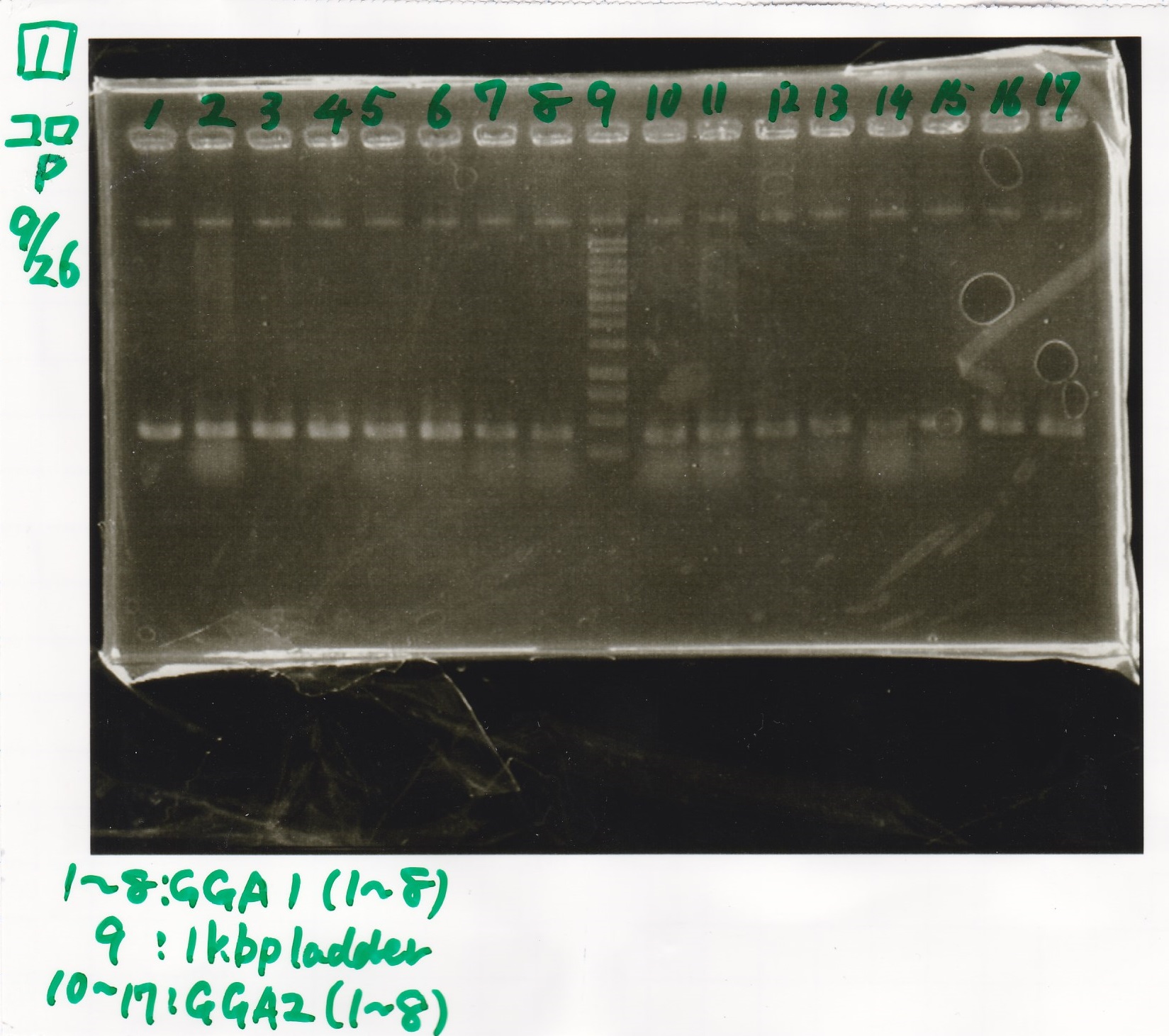
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | GGA3-1 | -- | --
|
| 2 | GGA3-2 | -- | --
|
| 3 | GGA3-3 | -- | --
|
| 4 | 1kbp ladder | -- | --
|
| 5 | GGA3-4 | -- | --
|
| 6 | GGA3-5 | -- | --
|
| 7 | GGA3-6 | -- | --
|
| 8 | GGA3-7 | -- | --
|
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | GGA3-8 | -- | --
|
| 2 | GGA4-1 | -- | --
|
| 3 | GGA4-2 | -- | --
|
| 4 | GGA4-3 | -- | --
|
| 5 | 1kbp ladder | -- | --
|
| 6 | GGA4-4 | -- | --
|
| 7 | GGA4-5 | -- | --
|
| 8 | GGA4-6 | -- | --
|
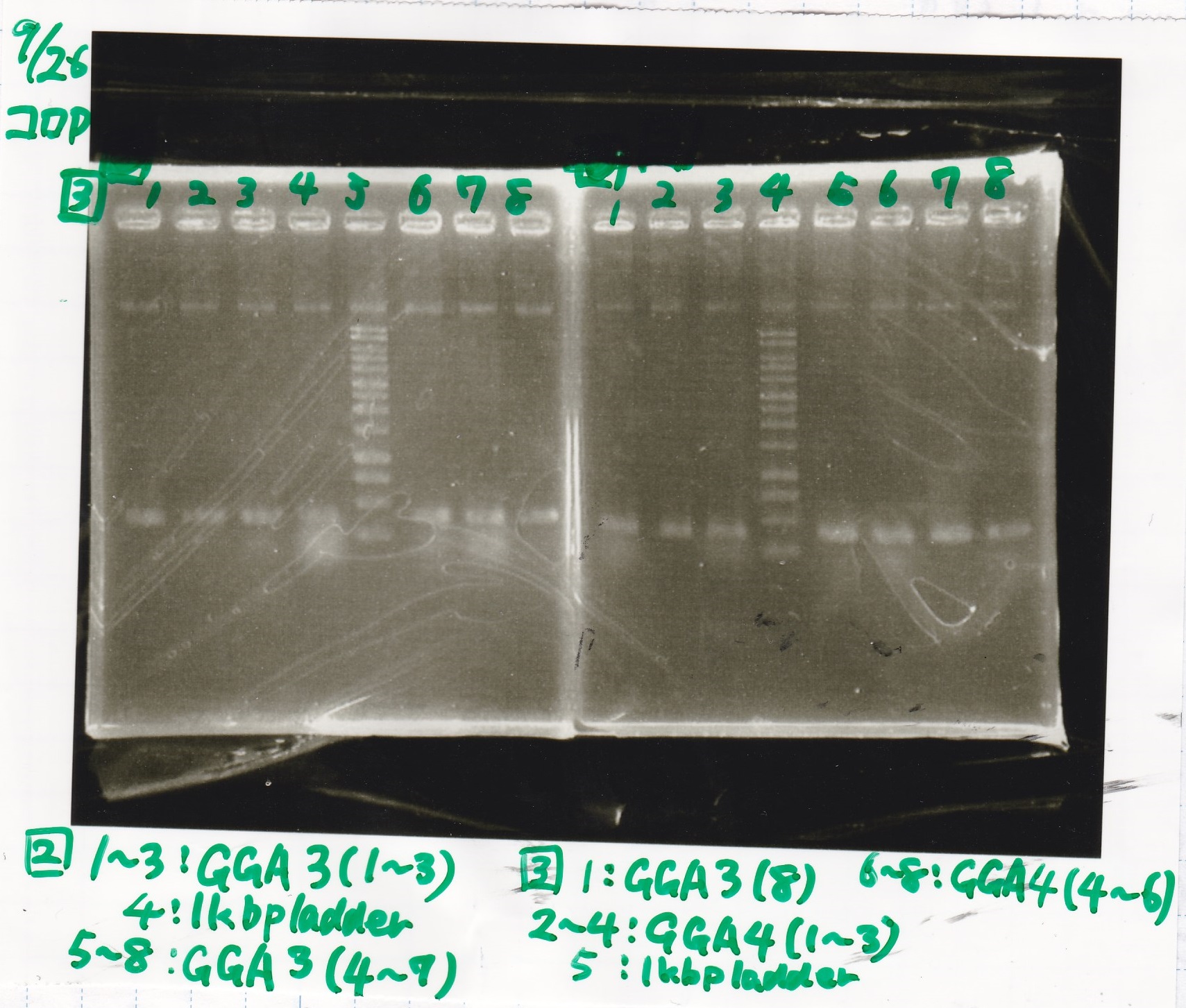
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | GGA4-7 | -- | --
|
| 2 | GGA4-8 | -- | --
|
| 3 | 1kbp ladder | -- | --
|
| 4 | GGA7-1 | -- | --
|
| 5 | GGA16-1 | -- | --
|
| 6 | NC(RBS-GFP-DT) | -- | --
|
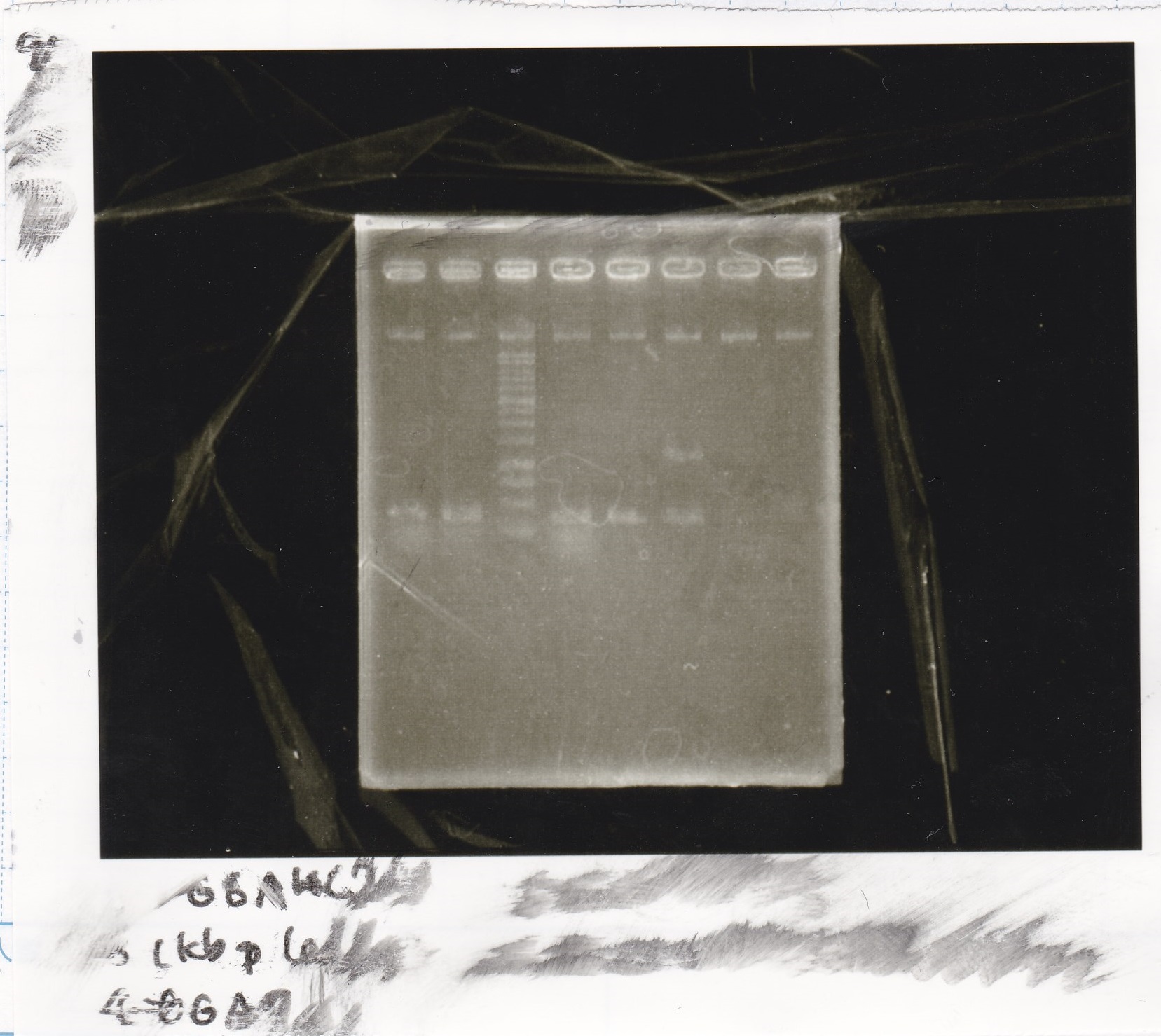
Ligasion(Golden Gate Assenbly)
No name
| state | Vector | Inserter1 | Inserter2 | Inserter3 | Inserter4 | Inserter5 | (NEB)T4 ligase | (NEB)10*T4 ligase | MilliQ
|
| experiment | RBS-GFP-DT | 1.0 µL | Pcon-pT181 antisense(E-2A) | 0.6µL | DT(2-1) | 0.5µL | Pcon-pT181 attenuator(1-SA) | 2.1 µL | -- | -- | -- | -- | 0.5 µL | 0.5 µL | up to 5 µL
|
| experiment | pSB1C3(EcoRI&SpeI) | 1.8 µL | Pcon-pT181 antisense(E-2A) | 0.9 µL | DT(2-1) | 0.5 µL | Pcon-GFP-DT(1-SA) | 1.5 µL | -- | -- | -- | -- | 0.5 µL | 0.5 µL | up to 5 µL
|
| experiment | RBS-GFP-DT | 1.0 µL | Ptet(E-0) | 0.7µL | Pcon pT181 attenuator(0-1A) | 0.3µL | tet aptamer(1-3) | 0.3 µL | Pcon-tetR-DT(3-2A) | 1.1 µL | Ptet(2-5) | 0.3 µL | 0.5 µL | 0.5 µL | up to 5 µL
|
| experiment | RBS-GFP-DT | 1.0 µL | Ptet(E-0) | 0.7 µL | Pcon-pT181 attenuator-DT(0-1A) | 1.0 µL | Pcon-tetR-DT(1-2A) | 1.0 µL | Ptet(2-5) | 0.3µL | -- | -- | 0.5 µL | 0.5 µL | up to 5 µL
|
| experiment | RBS-GFP-DT | 1.0 µL | Pcon-pT181 attenuator-tetR-DT(E-1A) | 1.0 µL | Pcon antisense-spinach-DT(1-2A) | 0.7µL | Pcon-pT 181 attenuator(2-SA) | 2.1 µL | -- | -- | -- | -- | 0.5 µL | 0.5 µL | up to 5 µL
|
| experiment | RBS-GFP-DT | 1.0 µL | Pcon antisense(E-1A) | 0.5 µL | spinach-DT(1-2) | 1.0 µL | Pcon-GFP-DT(1-2) | 0.8 µL | -- | -- | -- | -- | 0.5 µL | 0.5 µL | up to 5 µL
|
| experiment | pSB1C3(EcoRI&SpeI) | 1.8 µL | Pcon-pT181 antisense(E-1A) | 0.8µL | spinach-DT(1-2) | 1.0µL | Pcon-GFP-DT(2-SA) | 0.8 µL | -- | -- | -- | -- | 0.5 µL | 0.5 µL | up to 5 µL
|
| experiment | pSB1C3(EcoRI&SpeI) | 1.8 µL | Pcon-tet aptamer-DT(E-1A) | 3.0 µL | Ptet-pT181 antisense(1-3) | 1.5 µL | spinach-DT(3-2) | 1.5 µL | Pcon-GFP-DT(2-5A) | 0.8 µL | -- | -- | 0.5 µL | 0.5 µL | up to 5 µL
|
Liquid Culture
No name
| Sample | medium
|
| Pcon-GFP-DT | Amp
|
| RBS-GFP-DT | CP
|
| Pcon-pT181 attenuator-DT | Amp
|
| Pcon-tet aptamer-DT | Amp
|
| Pcon-spinach-DT | Amp
|
| Pcon-pT181 antisense-spinach-DT | Amp
|
| spinach-DT | CP
|
BSAI Digestion
No name
| | DNA | MilliQ | BSAI-HF | 10*cut smart | total
|
| Pcon | 7 µL | 10.7 µL | 0.3 µL | 2 µL | 20 µL
|
| RBS-luxR-DT | 10 µL | 7 µL | 0.3 µL | 2 µL | 20 µL
|
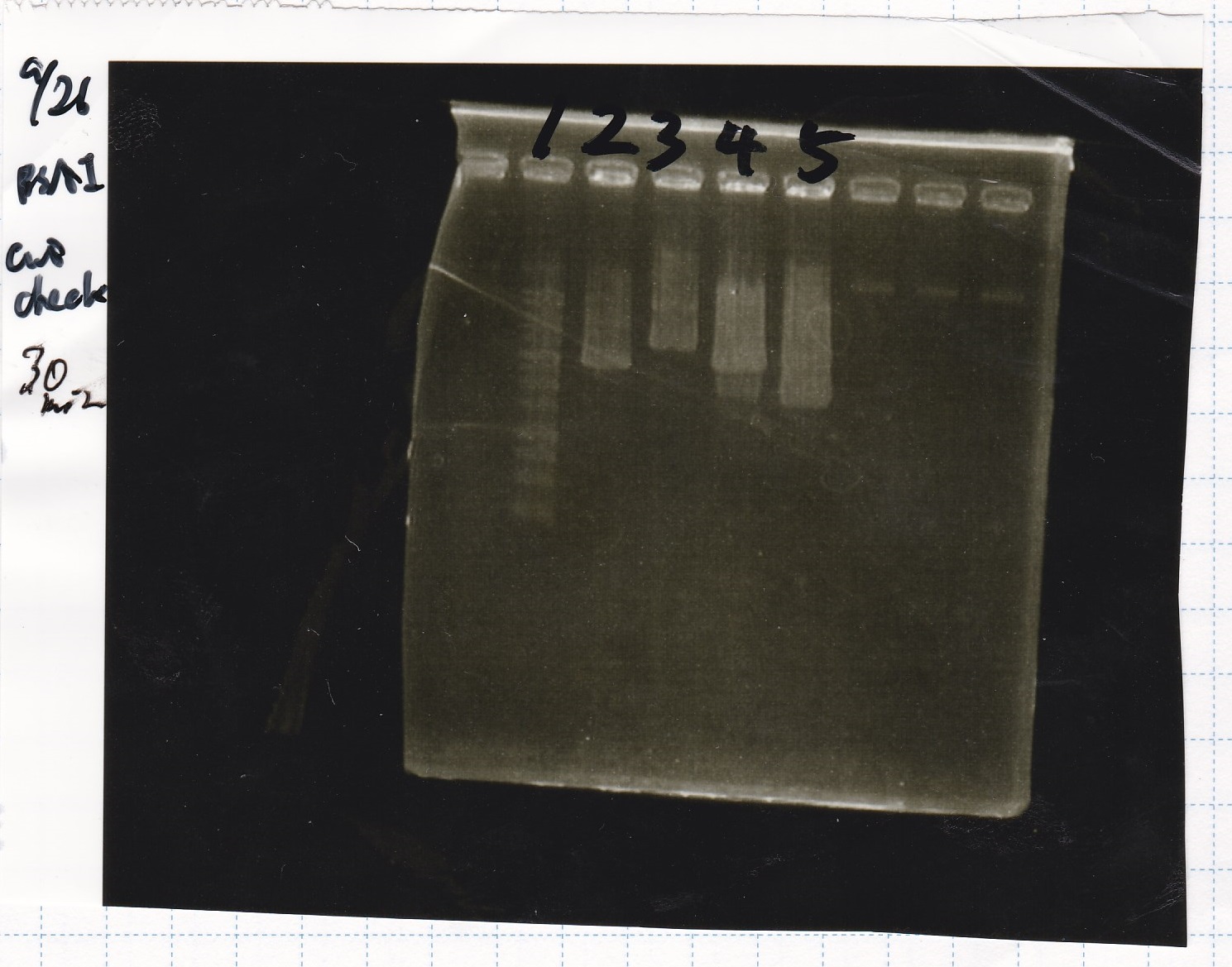
Colony PCR
Tatsui
| Sample | base pair
|
| GGA-1 (9~16) | 3177
|
| GGA-2 (9~16) | 3189
|
| GGA-3 (9~21) | 2989
|
| GGA-4 (9~16) | 2939
|
| GGA-6 (1) | 3359
|
| GGA-16(2~4) | 2939
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 3min30s | 30cycles
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | GGA-1 (9) | -- | --
|
| 2 | GGA-1 (10) | -- | --
|
| 3 | GGA-1 (11) | -- | --
|
| 4 | GGA-1 (12) | -- | --
|
| 5 | GGA-1 (13) | -- | --
|
| 6 | GGA-1 (14) | -- | --
|
| 7 | GGA-1 (15) | -- | --
|
| 8 | GGA-1 (16) | -- | --
|
| 9 | 1kbp ladder | -- | --
|
| 10 | GGA-2 (9) | -- | --
|
| 11 | GGA-2 (10) | -- | --
|
| 12 | GGA-2 (11) | -- | --
|
| 13 | GGA-2 (12) | -- | --
|
| 14 | GGA-2 (13) | -- | --
|
| 15 | GGA-2 (14) | -- | --
|
| 16 | GGA-2 (15) | -- | --
|
| 17 | GGA-2 (16) | -- | --
|
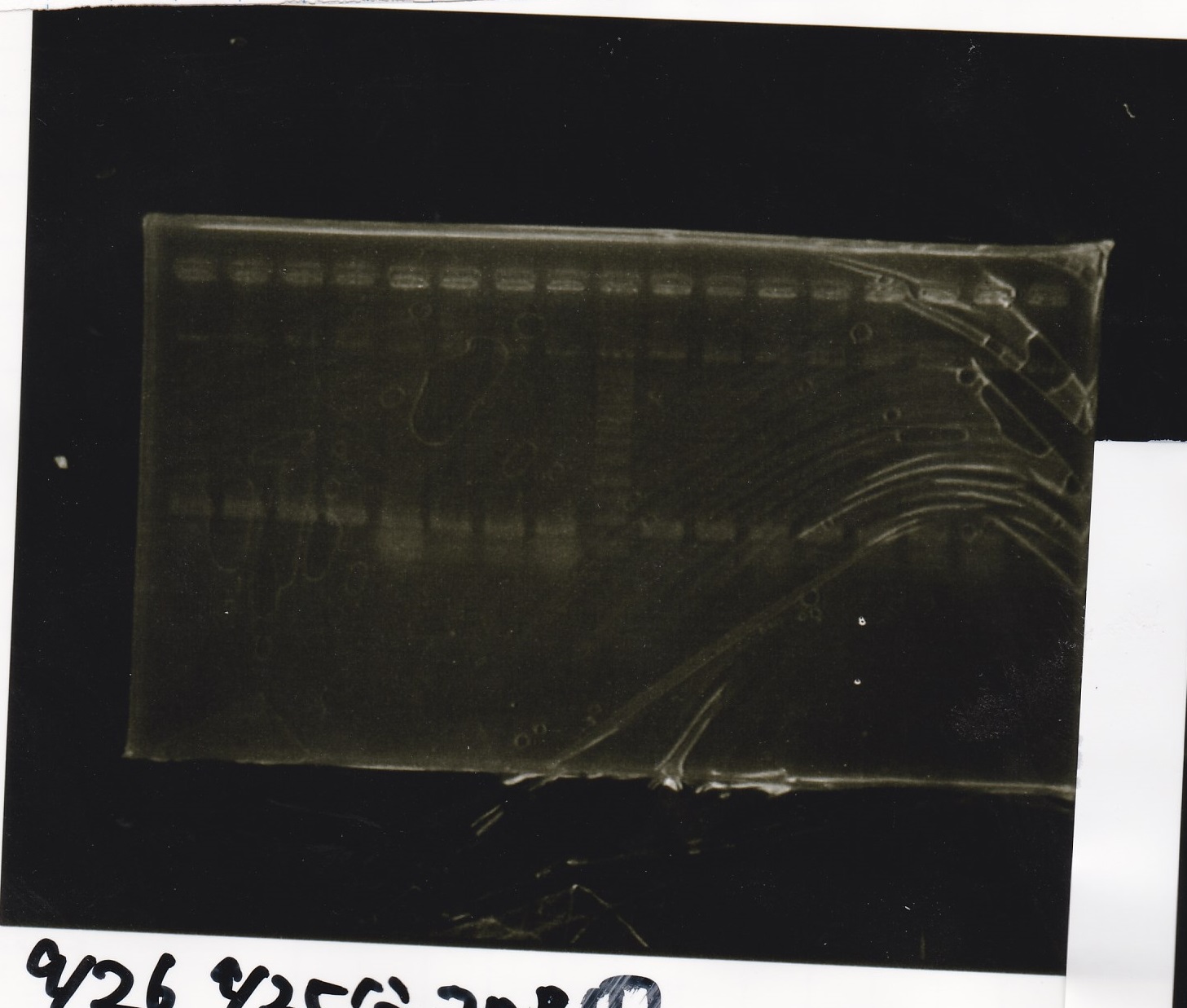
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | GGA-4 (9) | -- | --
|
| 2 | GGA-4 (10) | -- | --
|
| 3 | GGA-4 (11) | -- | --
|
| 4 | GGA-4 (12) | -- | --
|
| 5 | GGA-4 (13) | -- | --
|
| 6 | GGA-4 (14) | -- | --
|
| 7 | GGA-4 (15) | -- | --
|
| 8 | GGA-4 (16) | -- | --
|
| 9 | 1kbp ladder | -- | --
|
| 10 | GGA-6 (1) | -- | --
|
| 11 | GGA-16 (2) | -- | --
|
| 12 | GGA-16 (3) | -- | --
|
| 13 | GGA-16 (4) | -- | --
|
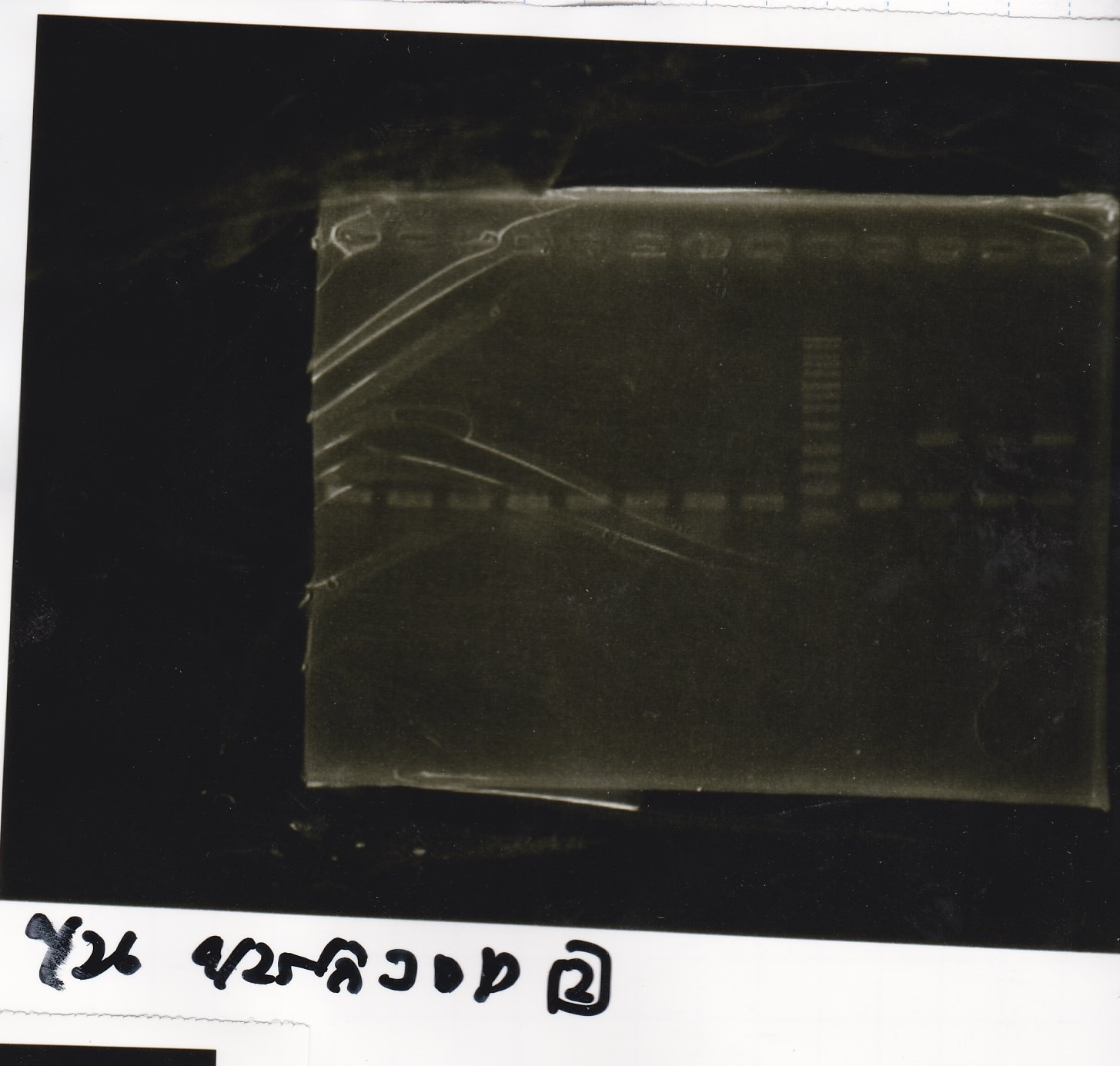
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | GGA-3 (9) | -- | --
|
| 2 | GGA-3 (10) | -- | --
|
| 3 | GGA-3 (11) | -- | --
|
| 4 | GGA-3 (12) | -- | --
|
| 5 | 1kbp ladder | -- | --
|
| 6 | GGA-3 (13) | -- | --
|
| 7 | GGA-3 (15) | -- | --
|
| 8 | GGA-3 (14) | -- | --
|
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | GGA-3 (17) | -- | --
|
| 2 | GGA-3 (18) | -- | --
|
| 3 | GGA-3 (19) | -- | --
|
| 4 | 1kbp ladder | -- | --
|
| 5 | GGA-3 (20) | -- | --
|
| 6 | GGA-3 (21) | -- | --
|
| 7 | GGA-3 (16) | -- | --
|
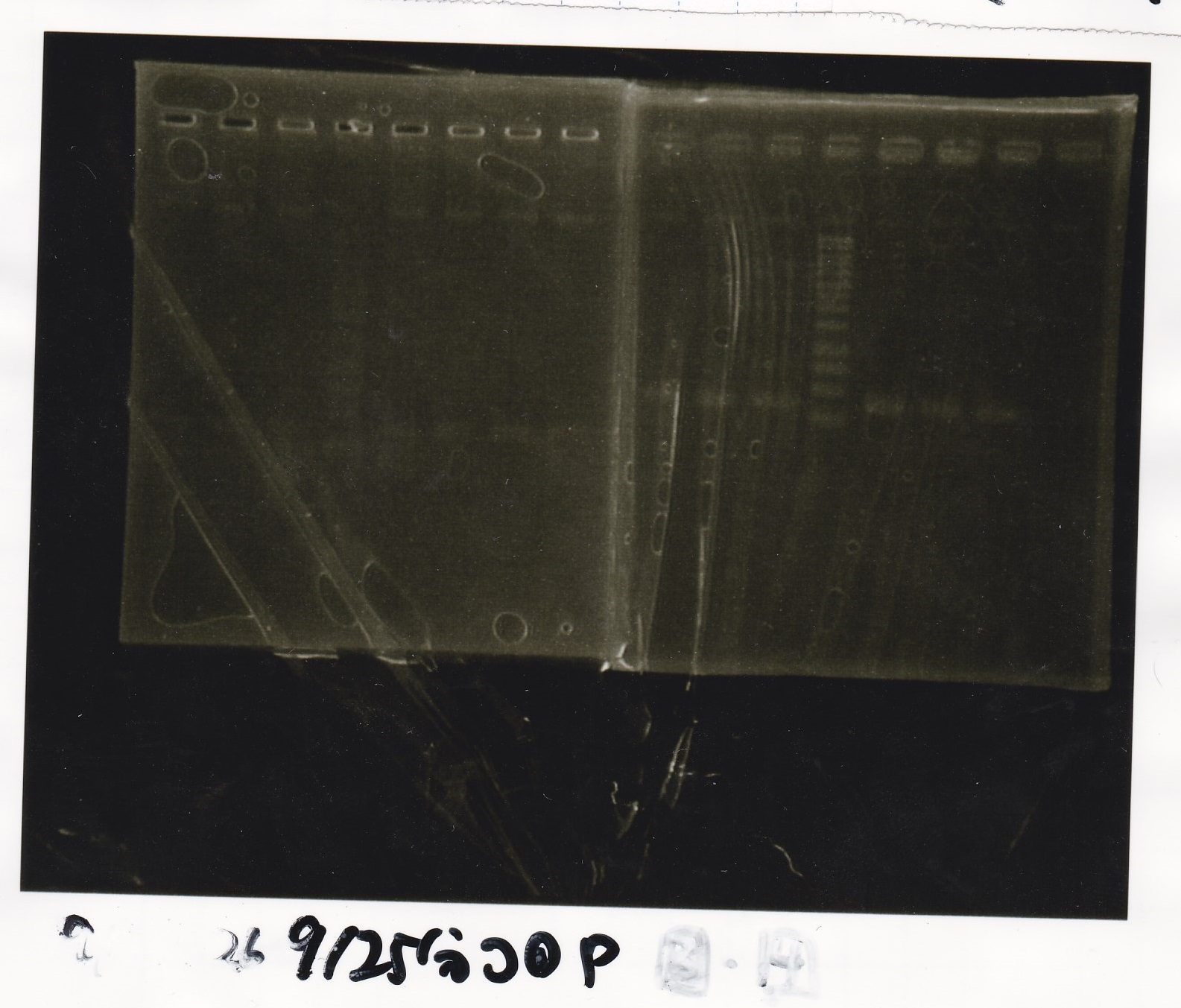
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | RBS-GFP-DT(RNA) | -- | --
|
| 2 | 1kbp ladder | -- | --
|
| 3 | Pcon-RBS-GFP-DT(RNA) | -- | --
|

Liquid Culture
Hirano
| Sample | medium
|
| Pcon-GFP-DT | --
|
| RBS-GFP-DT | --
|
| Pcon-tet aptamer-DT | --
|
| Pcon-spinach-DT | --
|
| Pcon-pT181 antisense-spinach-DT | --
|
| spinach-DT | --
|
Colony PCR
Hirano
| Sample | base pair
|
| 9/25 Pcon-pH181 attenuator+RBS-GFP-DT-1 | --
|
| 9/25 Pcon-pH181 attenuator+RBS-GFP-DT-2 | --
|
| 9/25 Pcon-pH181 attenuator+RBS-GFP-DT-3 | --
|
| 9/25 Pcon-pH181 attenuator+RBS-GFP-DT-4 | --
|
| 9/24 Ptet-RBS-GFP-DT-1 | --
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 68°C | --
|
| 5min | 30s | 30s | 1min40s | 30cycles
|
Ligasion(Golden Gate Assenbly)
Hirano
| state | Vector | Inserter1 | Inserter2 | Inserter3 | Inserter4 | Inserter5
|
| experiment | GGA2 | Pcon-attenuator(E-1A) | Pcon-tetR-DT(1-2A) | Ptet(2-S) | RBS-GFP-DT(EcoRI&XbaI) |
|
| experiment | GGA3 | Pcon-tetap-DT(E-1A) | Pcon-tetR-DT(1-2A) | Ptet(2-S) | RBS-GFP-DT(EcoRI&XbaI) |
|
| experiment | GGA4 | Pcon-spi-DT(E-1A) | Pcon-tetR-DT(1-2A) | Ptet(2-S) | RBS-GFP-DT(EcoRI&XbaI) |
|
| experiment | GGA5 | Pcon-antisense(E-1A) | DT(1-3) | Pcon-tetR-DT(3-2A) | Ptet(2-S) | RBS-GFP-DT(EcoRI&XbaI)
|
| experiment | GGA6 | Pcon-attenuator(E-1A) | tetap-DT(1-3) | Pcon-tetR-DT(3-2A) | Ptet(2-S) | RBS-GFP-DT(EcoRI&XbaI)
|
| experiment | GGA7 | Pcon-tetR-DT(E-2A) | Ptet(2-S) | RBS-GFP-DT(EcoRI&XbaI) | |
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 37°C | 16°C | 50°C | 80°C | --
|
| 3min | 4min | 5min | 5min | 25cycles
|
Nakamoto
| Sample
|
| Pcon-Spinach-DT
|
| Pcon-tetRaptamer-DT
|
| Pcon-attenuator-DT
|
| Spinach-DT
|
| Pcon-antisense-Spinach-DT
|
Liquid Culture
Kozima
| Sample
|
| Pcon-attemuater-DT
|
| Pcon-aptamer-DT
|
| Pcon-spinach-DT
|
| Pcon-RBS-tetR-DT
|
| Pcon-antisense-spinach-DT(Master 1)
|
| Pcon-antisense
|
Colony PCR
Nakamoto Kozima
| Sample | base pair
|
| 9/25 9/19pSB1C3&9/3PT181 attenuator(1~3) | 601bp
|
| 9/25 aptamer12_1R-DT&9/24Pcon-pT181 attenuator(1~4) | 859bp
|
| Pcon-attenuator-aptamer-DT(1) | 859bp
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 94°C | 55°C | 65°C | --
|
| 5min | 30sec | 30sec | 54sec | 30
|
Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | 1kbp ladder | -- | --
|
| 2 | 9/25 9/23 1C3&9/3attenuator | -- | --
|
| 3 | 9/25 9/23 1C3&9/3attenuator | -- | --
|
| 4 | 9/25 9/23 1C3&9/3attenuator | -- | --
|
| 5 | 9/25aptamer-DT&9/24Pcon-attenuator | -- | --
|
| 6 | 9/25aptamer-DT&9/24Pcon-attenuator | -- | --
|
| 7 | 9/25aptamer-DT&9/24Pcon-attenuator | -- | --
|
| 8 | 9/24 Pcon attenuator-aptamer-DT(1) | -- | --
|
| 9 | 9/25aptamer-DT&9/24Pcon-attenuator | -- | --
|
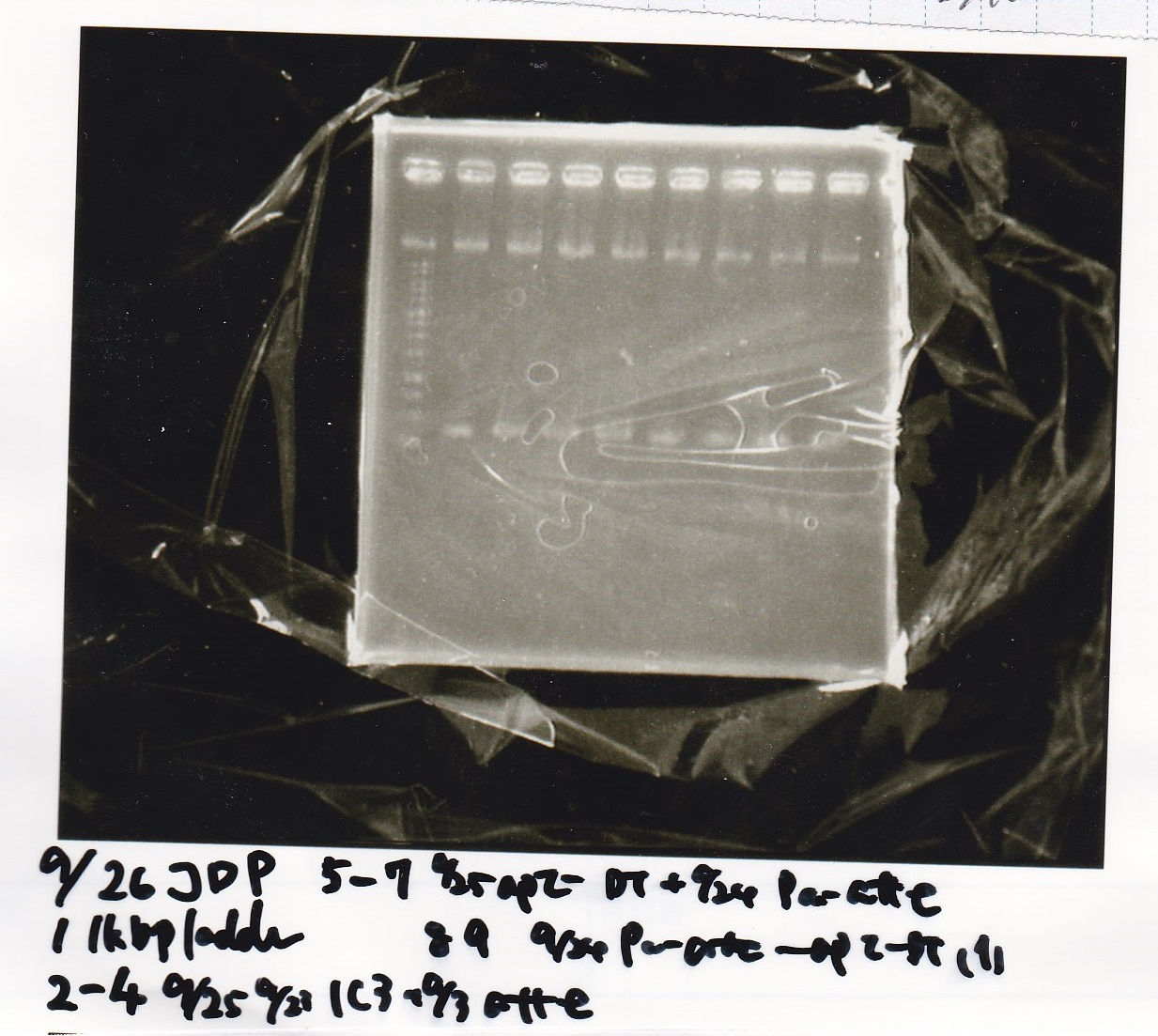
Plating
| Sample | Use plate
|
| 9/26 GGA2 | LB(CP)
|
| 9/26 GGA3 | LB(CP)
|
| 9/26 GGA4 | LB(CP)
|
| 9/26 GGA5 | LB(CP)
|
| 9/26 GGA6 | LB(CP)
|
| 9/26 GGA7 | LB(CP)
|
| 9/26 GGA11 | LB(CP)
|
| 9/26 GGA312 | LB(CP)
|
| 9/26 GGA13 | LB(CP)
|
| 9/26 GGA14 | LB(CP)
|
| 9/26 GGA15 | LB(CP)
|
| 9/26 GGA20 | LB(CP)
|
| 9/26 GGA23 | LB(CP)
|
| 9/26 GGA24 | LB(CP)
|
| 9/26 Ptet+RBS-GFP-DT | LB(CP)
|
incubate 37°C
Restriction Enzyme Digestion
Kojima
| 9/24 pSB1C3 | EcoRI | SpeI | buffer | MilliQ | total
|
| 2cuts | 11µL | 1µL | 1µL | 3µL | 14µL | 30µL
|
| NC | 0.5µL | 0µL | 0µL | 1µL | 8.5µL | 10µL
|
| 9/24 Pcon-pT181attenuator-aptamer12-1R-DT | EcoRI | SpeI | buffer | MilliQ | total
|
| 2cuts | 8.7µL | 1µL | 1µL | 3µL | 16.3µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 8.6µL | 10µL
|
| 9/24 pT181attenuator | EcoRI | SpeI | buffer | MilliQ | total
|
| 2cuts | 3.1µL | 1µL | 1µL | 3µL | 21.9µL | 30µL
|
| NC | 0.2µL | 0µL | 0µL | 1µL | 8.8µL | 10µL
|
| 8/17 RBS-GFP-DT | EcoRI | BSA | buffer | MilliQ | total
|
| 1cut | 16.4µL | 1µL | 3µL | 3µL | 6.6µL | 30µL
|
| NC | 0.4µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| 9/17 pSB4K5 | EcoRI | PstI | BSA | buffer | MilliQ | total
|
| 2cuts | 8.5µL | 1µL | 1µL | 3µL | 3µL | 13.5µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| 9/14 pSB4K5 | EcoRI | PstI | BSA | buffer | MilliQ | total
|
| 2cuts | 7.3µL | 1µL | 1µL | 3µL | 3µL | 14.7µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| 9/22 Pcon-pT181attenuator-DT | EcoRI | SpeI | buffer | MilliQ | total
|
| 2cuts | 5.4µL | 1µL | 1µL | 3µL | 19.6µL | 30µL
|
| NC | 0.3µL | 0µL | 0µL | 1µL | 8.7µL | 10µL
|
| 8/7 RBS-GFP-DT 1 | XbaI | PstI | BSA | buffer | MilliQ | total
|
| 2cuts | 8.1µL | 1µL | 1µL | 3µL | 3µL | 13.9µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
| 9/26 RBS-GFP-DT 1 | XbaI | BSA | buffer | MilliQ | total
|
| 1cut | 8.1µL | 1µL | 3µL | 3µL | 14.9µL | 30µL
|
| 9/16 Pλ-luxI | EcoRI | PstI | BSA | buffer | MilliQ | total
|
| 2cuts | 14.7µL | 1µL | 1µL | 3µL | 3µL | 7.3µL | 30µL
|
| NC | 0.7µL | 0µL | 0µL | 1µL | 1µL | 7.3µL | 10µL
|
| 9/21 Ptet | EcoRI | SpeI | buffer | MilliQ | total
|
| 2cuts | 14.2µL | 1µL | 1µL | 3µL | 10.8µL | 30µL
|
| NC | 0.7µL | 0µL | 0µL | 1µL | 8.3µL | 10µL
|
| 9/24 Pcon-pT181attenuator-aptamer12-1R-DT | XbaI | PstI | BSA | buffer | MilliQ | total
|
| 2cuts | 8.7µL | 1µL | 1µL | 3µL | 3µL | 13.3µL | 30µL
|
| NC | 0.4µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL
|
Electrophoresis
Kojima
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | pSB1C3 | EcoRI | SpeI
|
| 2 | pSB1C3 | -- | --
|
| 3 | Pcon-pT181attenuator-aptamer12-1R-DT | EcoRI | SpeI
|
| 4 | Pcon-pT181attenuator-aptamer12-1R-DT | -- | --
|
| 5 | 1kb ladder | -- | --
|
| 6 | pT181attenuator | EcoRI | SpeI
|
| 7 | pT181attenuator | -- | --
|
| 8 | RBS-GFP-DT | EcoRI | --
|
| 9 | RBS-GFP-DT | -- | --
|
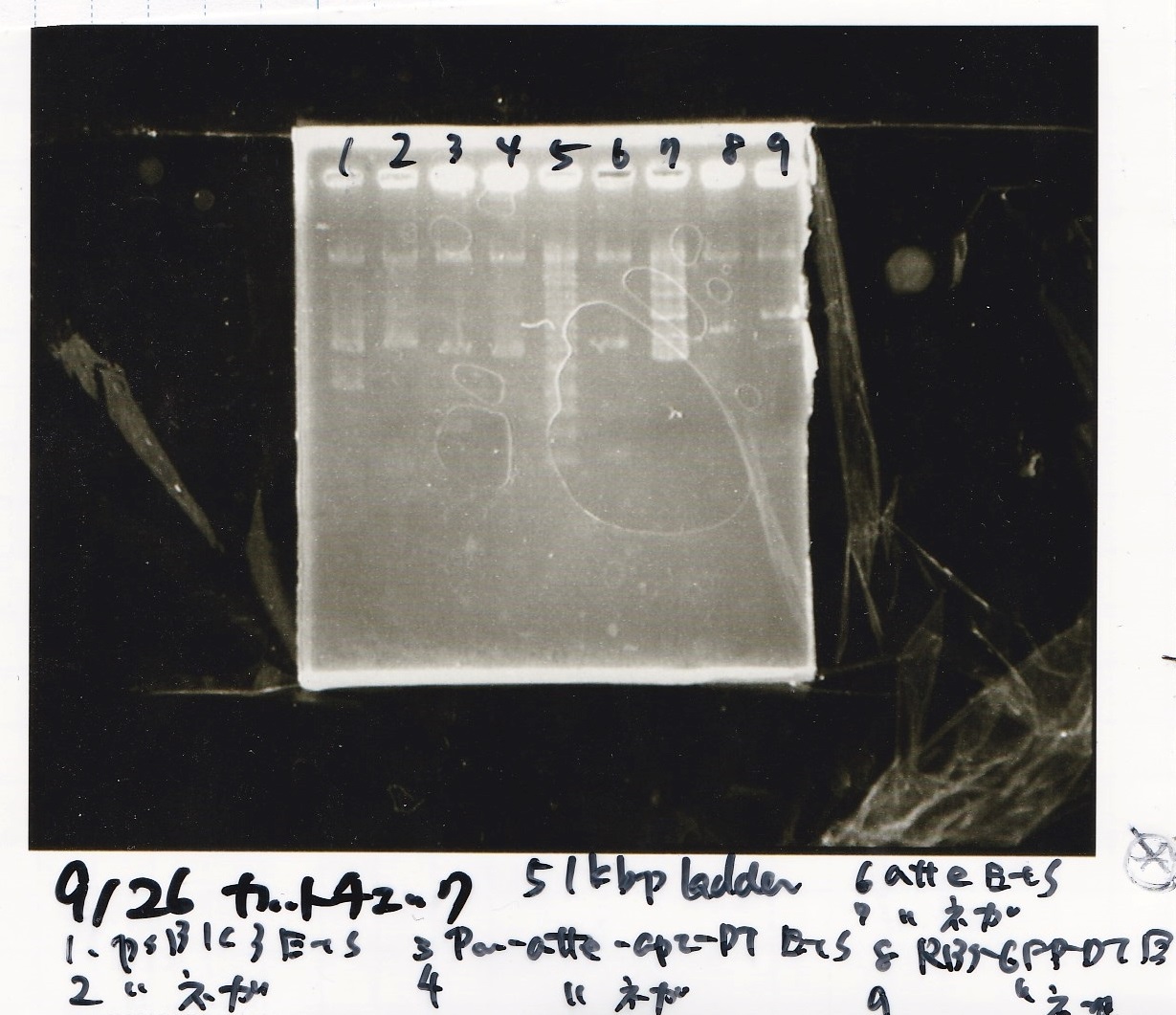
Sep 27
Kojima
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 2 | pSB1C3 | EcoRI+SpeI
|
| 3
|
| 4
|
| 6 | Pcon-attenuator-aptmer-DT | EcoRI+SpeI
|
| 7
|
| 8
|
| 10 | Pcon-attenuator-aptmer-DT | EcoRI+SpeI
|
| 11
|
| 12
|

| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-attenuator-DT(EcoRI+SpeI) | 6.8 | 1.98 | 0.43
|
| pT181-attenuator(EcoRI+SpeI) | 7.5 | 1.84 | 0.06
|
| pSB1C3(EcoRI+SpeI) | 17.7 | 1.97 | 0.71
|
No name
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 3 | pSB4K5 | EcoRI+PstI
|
| 4 | pSB4K5 | EcoRI+PstI
|
| 5 | pSB4K5 | EcoRI+PstI
|
| 7 | pSB6A1 | EcoRI+PstI
|
| 8 | pSB6A1 | EcoRI+PstI
|
| 9 | pSB6A1 | EcoRI+PstI
|

Electrophoresis
No name
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | pSB4K5 | EcoRI | PstI
|
| 2 | pSB4K5 | - | -
|
| 3 | 1kbp ladder | - | -
|
| 4 | Pcon-attenuator | EcoRI | SpeI
|
| 5 | Pcon-attenuator | - | -
|
| Lane | Sample | Enzyme1 | Enzyme2
|
| 1 | RBS-GFP-DT | XbaI | PstI
|
| 2 | RBS-GFP-DT | - | -
|
| 3 | RBS-GFP-DT | EcoRI | XbaI
|
| 4 | 1kbp ladder | - | -
|
| 5 | Ptet | EcoRI | SpeI
|
| 6 | Ptet | - | -
|

Liquid Culture
No name
| Sample | medium
|
| Pcon-GFP | Amp
|
| RBS-GFP-DT | CP
|
| Pcon-attenuator | Amp
|
| Pcon-attenuator-aptamer-DT | Amp
|
| Pcon-tetR-DT | Amp
|
Ligation
No name
| state | Vector | Inserter | Ligation High ver.2
|
| experiment | 9/17 pSB1C3 (EcoRI+SpeI) 10.0ng/µL | 10µL | 9/26 pT181 attenuator(EcoRI+SpeI) 7.5ng/µL | 1.3µL | 3.5 µL
|
| experiment | 9/25 DT (EcoRI+XbaI) 42.8ng/µL | 2.3µL | 9/22 Pcon-pT181 antisense(EcoRI+SpeI) 46.7ng/µL | 1.5µL | 1.9 µL
|
| experiment | 9/27 pSB4K5(EcoRI+PstI) 12.9ng/µL | 7.8µL | 9/10 Pcon-attenuator(EcoRI+SpeI) 13.8ng/µL | 8.0µL | 3.5 µL
|
| experiment | 9/27 pSB4K5(EcoRI+PstI) 12.9ng/µL | 7.8µL | 9/25 RBS-GFP-DT(XbaI+PstI) 13.9ng/µL | 12µL | 3.5 µL
|
| experiment | 9/8 pSB4K5 (EcoRI+SpeI) 17.6ng/µL | 5.7 µL | 8/21 Pcon-GFP-DT(EcoRI+SpeI) 11ng/µL | 12.6µL | 3.5 µL
|
Liquid Culture
Hirano
| Sample | medium
|
| Pcon-spinach-DT | plusgrow Amp
|
| Pcon-tetRaptamer-DT | plusgrow Amp
|
| Pcon-antisense-spinach-DT | plusgrow Amp
|
| spinach-DT | plusgrow CP
|
Nakamoto
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 9/26 Pcon-GFP-DT | 423.0 | 1.83 | 1.46
|
| 9/26 Pcon-GFP-DT K | 298.7 | 1.82 | 1.50
|
Electrophoresis
No name
| Lane | Sample
|
| 1 | 9/27 Pcon-GFP-DT(RNA)
|
| 2 | 1kb ladder
|
| 3 | 9/27 Pcon-GFP-DT K(RNA)
|

Miniprep
Tatsui
| DNA
|
| Pcon-aptamer-DT
|
| Pcon-spinach-DT
|
| Pcon-antisense
|
| Pcon-tetR-DT
|
| Pcon-attenuator-DT
|
| Pcon-antisense-spinach-DT
|
Transformation
Hirano
| Name1 | Name2 | Sample1(µL) | Sample2(µL) | Competent Cells(µL) | Total(µL)
|
| Pcon-attenuator+RBS-GFP-DT(4K5) | Pcon-attenuator-DT(1A2) | 2 | 1 | 30 | 33
|
| Pcon-attenuator+RBS-GFP-DT(4K5) | Pcon-antisense-spinach-DT(1A2) | 2 | 1 | 30 | 33
|
| Pcon-attenuator+RBS-GFP-DT(4K5) | Pcon-tetR aptamer-DT(1A2) | 2 | 1 | 30 | 33
|
| Pcon-RBS-GFP-DT(4K5) | Pcon-attenuator-DT(1A2) | 2 | 1 | 30 | 33
|
| Pcon-RBS-GFP-DT(4K5) | Pcon-antisense-spinach-DT(1A2) | 2 | 1 | 30 | 33
|
| Pcon-RBS-GFP-DT(4K5) | Pcon-tetR aptamer-DT(1A2) | 2 | 1 | 30 | 33
|
| Pcon-attenuator+RBS-GFP-DT(4K5) | -- | 2 | -- | 20 | 22
|
| Pcon-RBS-GFP-DT(4K5) | -- | 2 | -- | 20 | 22
|
| attenuator(1C3) | -- | 2 | -- | 20 | 22
|
Miniprep
No name
| DNA | concentration[µg/mL] | 260/280 | 260/230
|
| Pcon-RBS-GFP-DT | 137.6 | 1.95 | 1.78
|
| RBS-GFP-DT | 77.0 | 2.06 | 1.96
|
| Pcon-PT181 attenuator | 231.0 | 1.89 | 1.89
|
| Pcon-antisense-Spinach-DT(2) | 130.0 | 1.56 | 1.62
|
| Pcon-aptamer-DT | 218.8 | 1.92 | 1.72
|
| Pcon-attenuator-DT | 180.7 | 1.95 | 1.78
|
| Pcon-Spinach-DT | 177.6 | 1.96 | 1.06
|
| Pcon-antisense-Spinach-DT(1) | 181.1 | 1.96 | 1.92
|
| Pcon-antisense | 152.9 | 2.02 | 1.96
|
| Pcon-RBS+ezR-DT | 164.1 | 1.93 | 1.91
|
Restriction Enzyme Digestion
No name
| | 9/27 RBS-GFP-DT | XbaI | PstI | Buffer | BSA | MilliQ | total
|
| 2 cuts | 12µL | 1µL | 1µL | 3µL | 3µL | 20µL | 40µL
|
| NC | 1.3µL | 0µL | 0µL | 1µL | 1µL | 6.7µL | 10µL
|
| | 9/27 Pcon-tetR-DT | EcoRI | PstI | Buffer | MilliQ | total
|
| 2 cuts | 12.2µL | 1µL | 1µL | 3µL | 12.8µL | 30µL
|
| NC | 0.6µL | 0µL | 0µL | 1µL | 8.4µL | 10µL
|
Nakamoto
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| 9/27 Pcon-RBS-GFP-DT | 686.6 | 1.99 | 1.34
|
| 9/27 Pcon-RBS-GFP-DT (+High salt) | 127.6 | 1.76 | 0.27
|
DNA sythesis
No name
| genome DNA | DNA volume | genome DNA wipeout buffer | RNase freewater | RT | RT buffer | Primer Mix | total
|
| 9/27 Pcon-RBS-GFP-DT | 0.8µL | 1µL | 5.2µL | 0.5µL | 2µL | 0.5µL | 10µL
|
| 9/27 pcon-RBS-GFP-DT(+high salt) | 2.5µL | 1µL | 3.5µL | 0.5µL | 2µL | 0.5µL | 10µL
|
| 9/27 Pcon-RBS-GFP-DT(revised concentration) | 0.5µL | 1µL | 5.5µL | 0.5µL | 2µL | 0.5µL | 10µL
|
Electrophoresis
No name
| Lane | Sample
|
| 1 | 1kb ladder
|
| 2 | Pcon-RBS-GFP-DT
|
| 3 | Pcon-RBS-GFP-DT
|
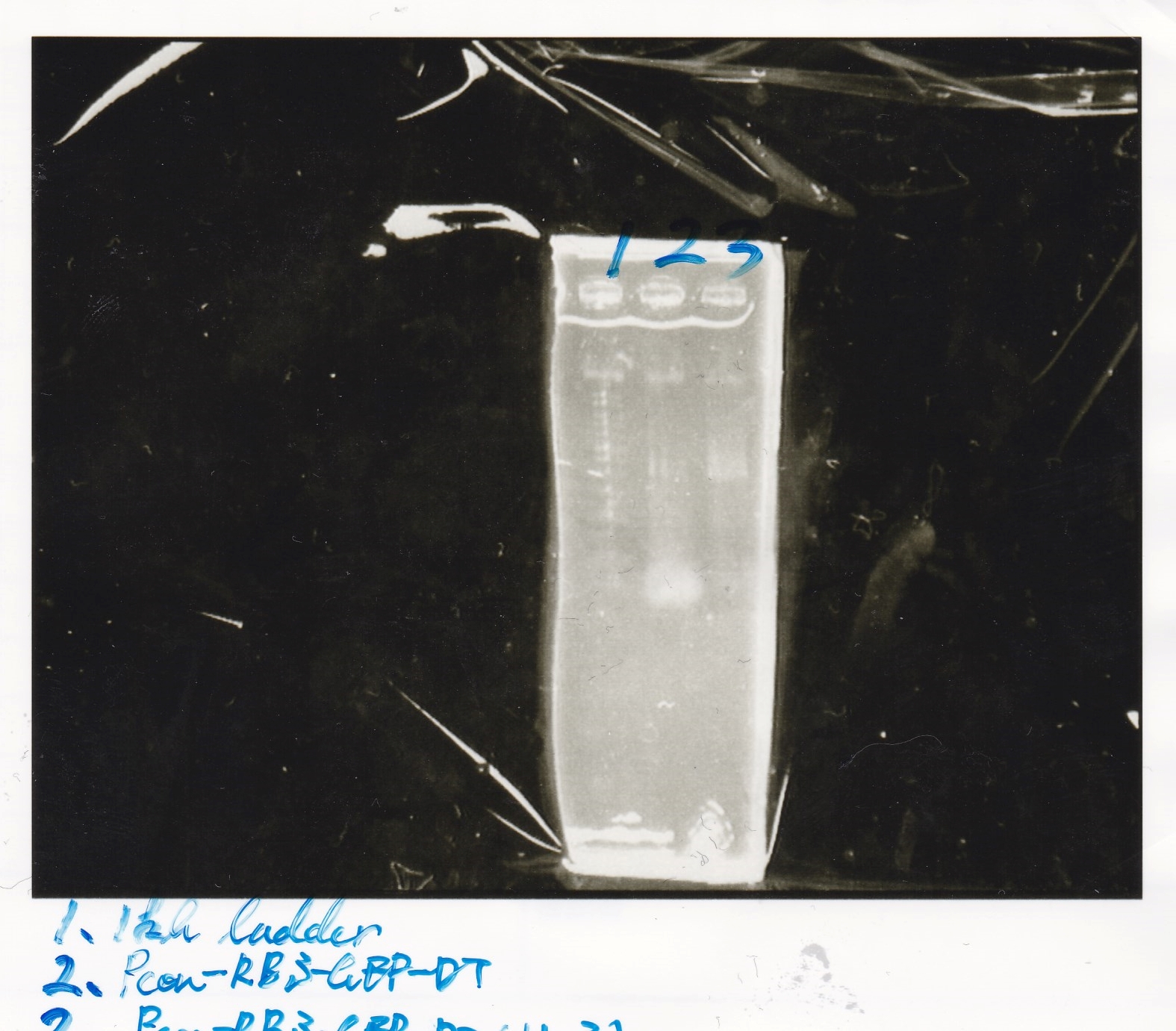
Liquid Culture
No name
| Sample | medium
|
| Pcon spinach DT | plusgrow 2µL(Amp)
|
| Pcon aptamer DT | plusgrow 2µL(Amp)
|
| Pcon antisense Spinach DT | plusgrow 2µL(Amp)
|
| Spinach DT | plusgrow 4µL(CP)
|
RT PCR
No name
| genome DNA | KOD plus | 10x buffer for KOD-plus-ver.2 | 2nM dNTPs | 25mM MgSO4 | F primer | R primer | MilliQ | total
|
| 0.5 | 1 | 2.5 | 2.5 | 1.5 | 0.75 | 0.75 | 15.5 | 25
|
| PreDenature | Denature | Annealing | Extension | cycle
|
| 94°C | 98°C | 49.8°C | 68°C | --
|
| 2min | 10sec | 30sec | 10sec | 30
|
Electrophoresis
No name
| Lane | Sample
|
| 1 | Pcon-GFP-DT(GFP)
|
| 2 | Pcon-GFP-DT(internal)
|
| 3 | Pcon-GFP-DT(HS)(GFP)
|
| 4 | Pcon-GFP-DT(internal)
|
| Lane | Sample
|
| 1 | Pcon-GFP-DT(GFP)
|
| 2 | Pcon-GFP-DT(internal)
|
| 3 | 100bp ladder
|

Digesting cut check
No name
| Lane | Sample
|
| 1 | RBS-GFP-DT(XbaI&PstI)
|
| 2 | RBS-GFP-DT NC(XbaI&PstI)
|
| 3 | 1kbp ladder
|
| 4 | Pcon-RBS-tetR-DT(EcoRI&SpeI)
|
| 5 | Pcon-RBS-tetR-DT NC(EcoRI&SpeI)
|

No name
| Lane | DNA | Enzyme
|
| 1 | 1kbp ladder | --
|
| 3 | RBS-GFP-DT | Xbal+PstI
|
| 4 | RBS-GFP-DT | Xbal+PstI
|
| 5 | RBS-GFP-DT | Xbal+PstI
|
| 7 | 1kbp ladder | --
|
| 9 | Pcon-tetR-DT | EcoRI+SpeI
|
| 10 | Pcon-tetR-DT | EcoRI+SpeI
|
| 11 | Pcon-tetR-DT | EcoRI+SpeI
|
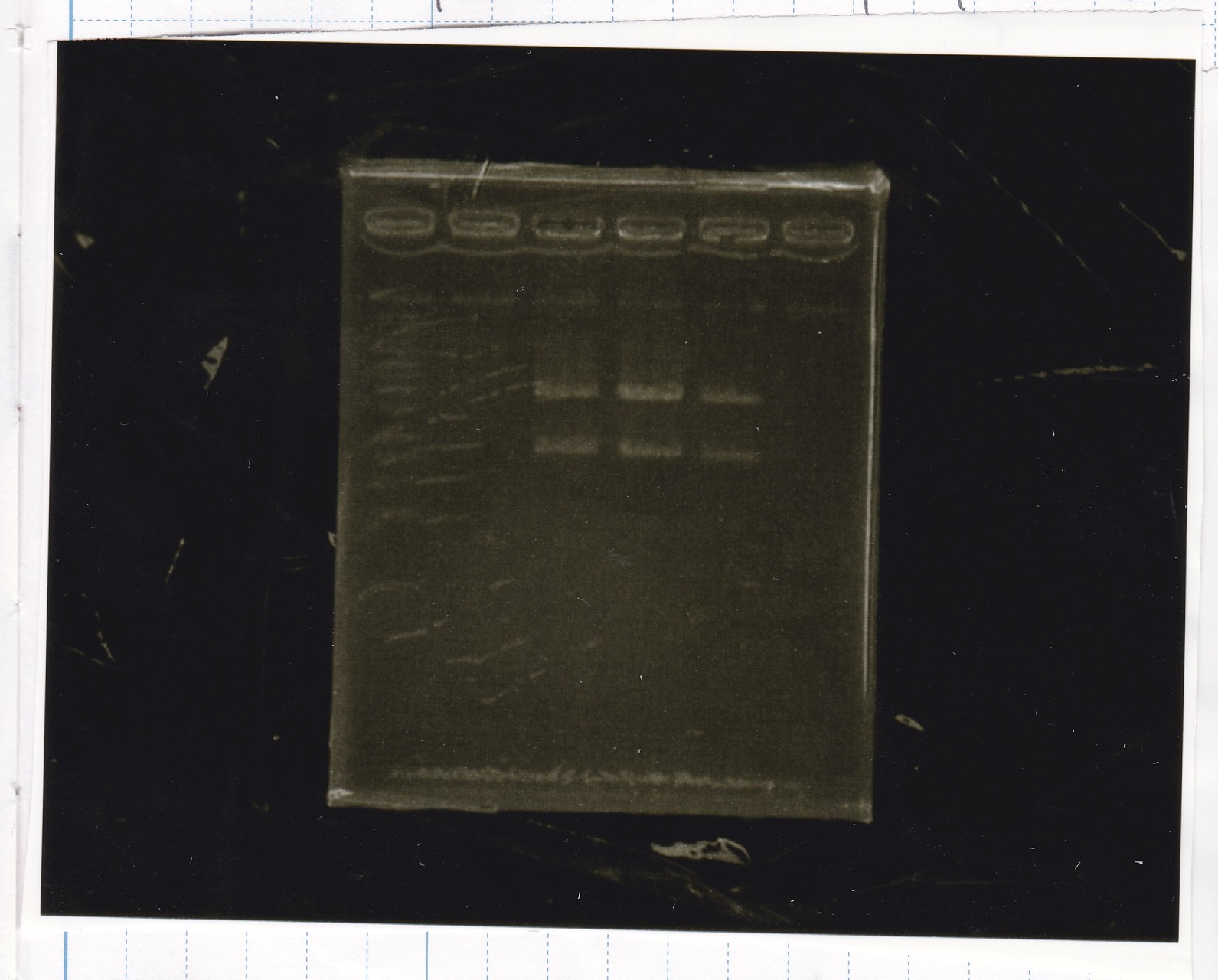

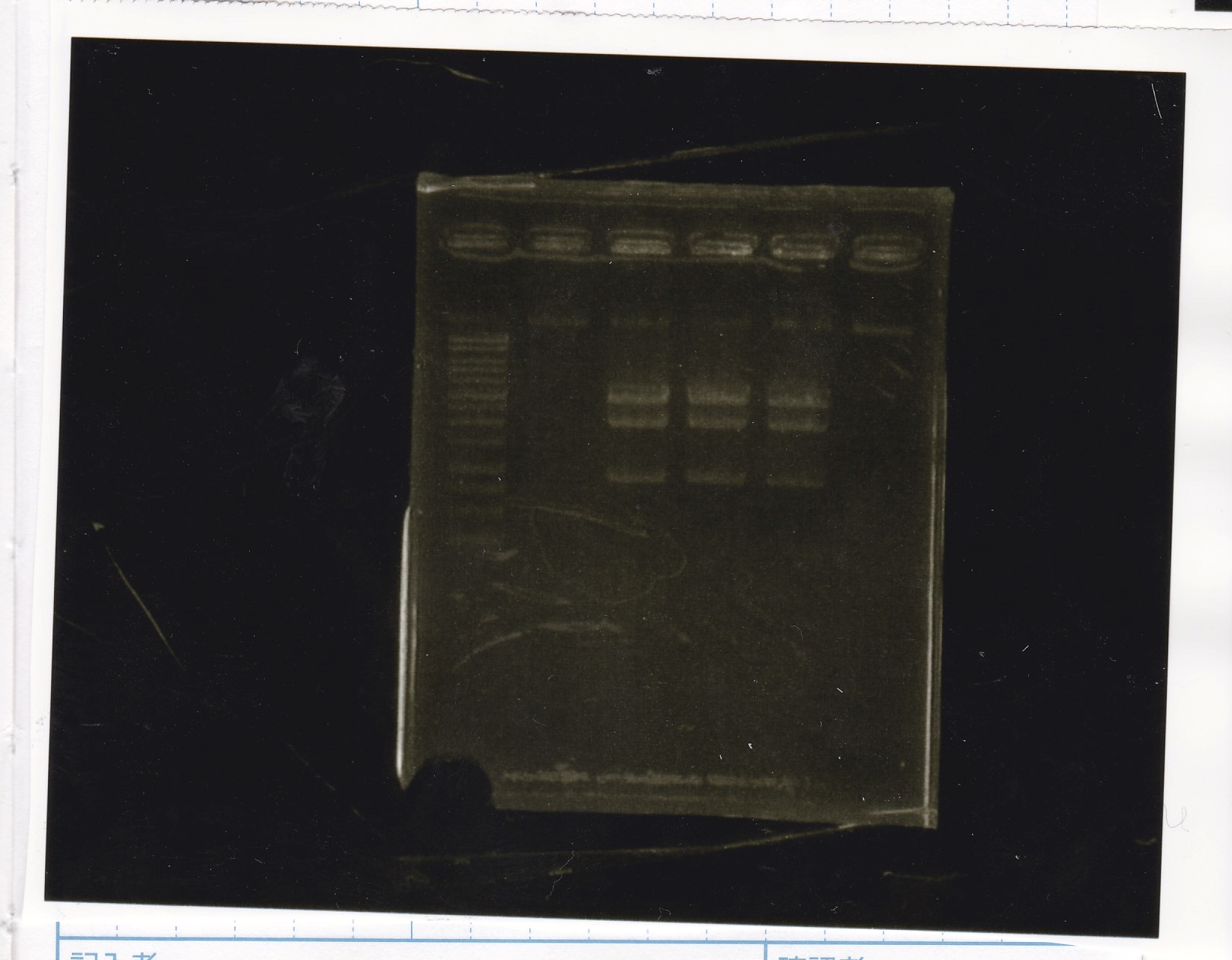

Nakamoto
| 0 | 5min | 15min | 20min | 23min | 28min | 33min | 36min
|
| aptamer generator | 0.348 | 0.408 | 0.550 | 0.865 | | | |
|
| Pcon-spinach-DT | 0.250 | 0.240 | 0.305 | 0.426 | 0.471 | 0.554 | 0.668 | 0
|
| GFP generator | 0.389 | 0.420 | 0.516 | 0.884 | | | |
|
| antisense-spinach | 0.371 | 0.438 | 0.537 | 0.733 | | | |
|
| Spinach-DT | 0.084 | 0.110 | 0.134 | 0.433 | 0.269 | 0.326 | 0.409 | 0.742
|
| Name | concentration[µg/mL] | 260/280 | 260/230
|
| Spinach-DT | 83.9 | 1.63 | 0.17
|
| Pcon-tetRaptamer-DT | 425.1 | 2.10 | 0.64
|
| Pcon-RBS-GFP-DT | 262.4 | 2.01 | 0.84
|
| Pcon-antisense-Spinach-DT | 489.1 | 2.05 | 0.68
|
| Pcon-Spinach-DT | 266.6 | 2.01 | 0.66
|
cDNA Synthesis
Nakamoto
| 9/27 Spinach-DT | 9/23 Pcon-tetRaptamer-DT | 9/23 Pcon-RBS-GFP-DT | 9/23 Pcon-antisense-spinach-DT | 9/23 Pcon-Spinach-DT
|
| RNA | 1 | 1 | 1 | 1 | 1
|
| gDNA wipeout | 6 | 1.2 | 1.9 | 1.0 | 1.9
|
| RNase-free water | 0 | 4.8 | 4.1 | 5.0 | 4.1
|
| RT | 0.5 | 0.5 | 0.5 | 0.5 | 0.5
|
| RT Buffer | 2 | 2 | 2 | 2 | 2
|
| Primer mix | 0.5 | 0.5 | 0.5 | 0.5 | 0.5
|
| total | 10µL | 10µL | 10µL | 10µL | 10µL
|
|
|
|
 "
"