Template:Kyoto/Notebook/Sep 11
From 2013.igem.org
Contents |
Sep 11
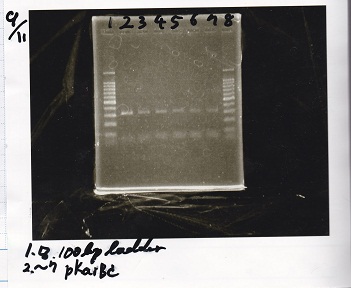
Electrophoresis
| Lane | Sample |
|---|---|
| 1 | 1kladder |
| 2 | Pbad/araC+RBS-luxI-DT-1 |
| 3 | Pbad/araC+RBS-luxI-DT-1 |
| 4 | Plac+RBS-lysis-DT |
| 5 | Pcon+RBS-luxI-DT |
| 6 | 1kladder |
PCR
| pKaiBC(200p) | 10*buffer KOD plus | dNTPs | MgSO4 | primer mix(F&R) | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 0.5 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 16 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 60°C | 68°C | 30 |
| 2min | 10sec | 30sec | 30sec |
| pKaiBC(400p) | 10*buffer KOD plus | dNTPs | MgSO4 | primer mix(F&R) | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 1 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 15.5 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 60°C | 68°C | 30 |
| 2min | 10sec | 30sec | 30sec |
| pKaiBC(800p) | 10*buffer KOD plus | dNTPs | MgSO4 | primer mix(F&R) | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 2 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 14.5 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 60°C | 68°C | 30 |
| 2min | 10sec | 30sec | 30sec |
| pKaiBC(1ng) | 10*buffer KOD plus | dNTPs | MgSO4 | primer mix(F&R) | KOD plus | MilliQ | total |
|---|---|---|---|---|---|---|---|
| 2.5 | 2.5 | 2.5 | 1.5 | 1.5 | 0.5 | 14 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 98°C | 60°C | 68°C | 30 |
| 2min | 10sec | 30sec | 30sec |
Electrophoresis
| Lane | Sample |
|---|---|
| 1 | 100bp ladder |
| 2 | pKaiBC 800pg 60°C |
| 3 | pKaiBC 1ng 60°C |
| 4 | pKaiBC 800pg 61°C |
| 5 | pKaiBC 1ng 61°C |
| 6 | pKaiBC 800pg 62°C |
| 7 | pKaiBC 1ng 62°C |
| 8 | 100bp ladder |
colony PCR
| Sample | base pair |
|---|---|
| 9/9Pcon-lacZα(p3B4k5) | 712bp |
| 9/9Pcon-spinach-DT(p3B4k5) | 605bp |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 42s |
| Sample | base pair |
|---|---|
| Pbad\araC-RBS-luxI-DT |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 5min | 30s | 30s | 2min |
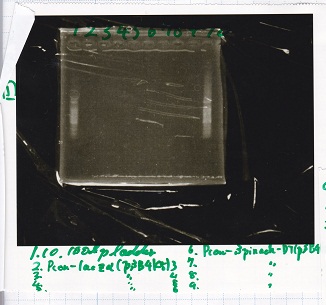
Electrophoresis
| Lane | Sample |
|---|---|
| 1 | 100bp ladder |
| 2 | Pcon-lacZα(pSB4K5)3 |
| 3 | Pcon-lacZα(pSB4K5)4 |
| 4 | Pcon-lacZα(pSB4K5)5 |
| 5 | Pcon-lacZα(pSB4K5)6 |
| 6 | Pcon-spinach-DT(pSB4K5)3 |
| 7 | Pcon-spinach-DT(pSB4K5)4 |
| 8 | Pcon-spinach-DT(pSB4K5)5 |
| 9 | Pcon-spinach-DT(pSB4K5)6 |
| 10 | 100bp ladder |
| Lane | Sample |
|---|---|
| 1 | 1kbp ladder |
| 2 | Pbad/araC-RBS-luxI-DT3 |
| 3 | Pbad/araC-RBS-luxI-DT4 |
| 4 | Pbad/araC-RBS-luxI-DT5 |
| 5 | Pbad/araC-RBS-luxI-DT6 |
| 6 | 1kbp ladder |
colony PCR
| Sample | base pair |
|---|---|
| 9/9Pcon+RBS-luxI-DT | 1079bp |
| 9/10Pcon+sptamer12_1R-DT | 584bp |
| 9/10DT+Pcon-pT181 attenuator 1 | 781bp |
| 9/10Fusion1 attenuator(p3B1C3)-1 | 638bp |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 55°C | 68°C | -- |
| 50sec | 30sec | 30sec | 1min |
Gel Extraction
Ligation
| state | Vector | Inserter | Ligation High ver.2 | ||
|---|---|---|---|---|---|
| experiment | 9/8pSB4K5(EcoRI&SpeI)21.8ng/µL | 2.3 µL | 9/8Pcon-Spinach-DT(EcoRI&SpeI)10.5ng/µL | 1.7 µL | 2.0 µL |
| experment | 9/8pSB4K5(EcoRI&SpeI)21.8ng/µL | 2.3 µL | 9/8RBS-lacZα-DT(EcoRI&SpeI)4.4ng/µL | 6.7µL | 3.5 µL |
| experiment | 9/8pSB4K5(EcoRI&SpeI)21.8ng/µL | 2.3 µL | 9/8Pcon-RBS-lacZα-DT(EcoRI&SpeI)5.6ng/µL | 1.9 µL | 2.1 µL |
| experiment | 9/6Plux 25ng/µL | 2 µL | 9/8 RBS-lysis1-DT(XbaI&PstI) | 6.6 µL | 3.5 µL |
| experiment | 9/6Plux 25ng/µL | 2 µL | 9/8 RBS-lysis3-DT(XbaI&PstI) | 4.7 µL | 3.4 µL |
| experiment | 9/10Pcon-pT181 attenuator(SpeI&PstI) | 3.4µL | 9/8RBS-lacZα-DT(EcoRI&SpeI) | 4.5 µL | 3.5 µL |
| experiment | 9/6Pcon-RBS laxR-DT(EcoRI&XbaI) 26.7ng/µL | 1.9 µL | Plax-RBS-GFP-DT | 5.5 µL | 3.5 µL |
| experiment | 9/9 Pcon-pT181 attenuator(SpeI&PstI) | 3.4 µL | 9/10 aptamer12_1R-DT(XbaI&PstI) | 1.7 µL | 2.6 µL |
Transformation
| Name | Sample | Competent Cells | Total | Plate | |
|---|---|---|---|---|---|
| 9/9 Fusion1 antisense(pSB2C3) | 2µL | 20µL | 22µL | CP | |
| 9/9 Fusion6 antisense(pSB2C3) | 2µL | 20µL | 22µL | CP | |
| 9/9 aptamer 12_P(pSB2C3) | 2µL | 20µL | 22µL | CP | |
| 9/9 aptamer 12_1µ(pSB2C3) | 2µL | 20µL | 22µL | CP | |
| 9/9 Fusion2 attenuator(pSB2C3) | 2µL | 20µL | 22µL | CP | |
| 9/11 Pcon-spinach-DT(pSB4K5) | 2µL | 20µL | 22µL | K | |
| 9/11 RBS-lacZα-DT(pSB4K5) | 2µL | 20µL | 22µL | K | |
| 9/11 Pcon-RBS-lacZα-DT(pSB4K5) | 2µL | 20µL | 22µL | K | |
| Plux+RBS-lysis1-DT | 2µL | 20µL | 22µL | CP | |
| Plux+RBS-lysis3-DT | 2µL | 20µL | 22µL | CP | |
| Pcon-pT181attenuator+RBS-lacZα-DT | 2µL | 20µL | 22µL | Amp | |
| Pcon-iuxR+Plux-GFP | 2µL | 20µL | 22µL | Amp | |
| Pcon-pT181attenuator+apt12_1R-DT | 2µL | 20µL | 22µL | Amp | |
| Plac(BBa_R0011) | 2µL | 20µL | 22µL | Amp |
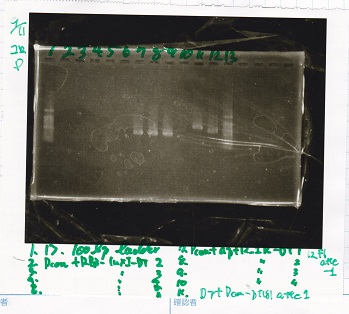
Electrophoresis
| Lane | Sample |
|---|---|
| 1 | 100bp ladder |
| 2 | Pcon+RBS luxI-DT2 |
| 3 | Pcon+RBS luxI-DT3 |
| 4 | Pcon+RBS luxI-DT4 |
| 5 | Pcon+RBS luxI-DT5 |
| 6 | Pcon+RBS luxI-DT6 |
| 7 | Pcon+apF12_1R-DT1 |
| 8 | Pcon+apF12_1R-DT2 |
| 9 | Pcon+apF12_1R-DT3 |
| 10 | Pcon+apF12_1R-DT4 |
| 11 | DT+Pcon-pT181attenuator1 |
| 12 | F1attenuator(pSB1C3) |
| 13 | 100bp ladder |
Restriction Enzyme Digestion
| DNA | EcoRI | XbaI | SpeI | PstI | buffer | BSA | MilliQ | Total | ||
|---|---|---|---|---|---|---|---|---|---|---|
| pSB4K5(EcoRI+SpeI) | 9.4µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 12.6µL | 30 µL | |
| pSB4K5(EcoRI+SpeI) negative | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL | |
| RBS-lacZα-DT(EcoRI+SpeI) | 10µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 12µL | 30µL | |
| RBS-lacZα-DT(EcoRI+SpeI) negative | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL | |
| Ptet-pT181 antisense(SteI+PstI) | 10.8µL | 0µL | 0µL | 1µL | 1µL | 3µL | 3µL | 12µL | 30µL | |
| Ptet-pT181 antisense(SteI+PstI) negative | 0.5µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.5µL | 10µL | |
| Pcon-RBS-lacZα-DT(EcoRI+SpeI) | 7.1µL | 1µL | 0µL | 1µL | 0µL | 3µL | 3µL | 14.9µL | 30µL | |
| Pcon-RBS-lacZα-DT(EcoRI+SpeI) negative | 0.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 7.6µL | 10µL | |
| RBS-lysis2-DT(XbaI+PstI) | 22µL | 0µL | 1µL | 0µL | 1µL | 3µL | 3µL | 0µL | 30µL | |
| RBS-lysis2-DT(XbaI+PstI) negative | 1.4µL | 0µL | 0µL | 0µL | 0µL | 1µL | 1µL | 6.6µL | 10µL |
Liquid Culture
| Sample | medium |
|---|---|
| 8/16 Master4 Plax | |
| 8/25 Pλ-laxI-2 |
PCR
| 8/29 Pλ-RBS-luxI-DT(76.5ng/µL) | 10x buffer for KOD plus-ver.2 | KOD-plus | dNTP | MgSO4 | F primer(RBS-luxI-DT-cloning-fwd) | R primer(RBS-luxI-DT-cloning-rev) | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.3 | 2.5 | 0.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.2 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 65°C | 68°C | -- |
| 5min | 30sec | 30sec | 1min | 30 |
| 8/31 Pbad/araC-RBS-RFP(1)(67.6ng/µL) | 10x buffer for KOD plus-ver.2 | KOD-plus | dNTP | MgSO4 | F primer(Pbad/araC(pSBlc3)-cloning-fwd) | R primer(Pbad/araC(pSBlc3)-cloning-rev) | MilliQ | total |
|---|---|---|---|---|---|---|---|---|
| 0.3 | 2.5 | 0.5 | 2.5 | 1.5 | 0.75 | 0.75 | 16.2 | 25 |
| PreDenature | Denature | Annealing | Extension | cycle |
|---|---|---|---|---|
| 94°C | 94°C | 54°C | 68°C | -- |
| 5min | 30sec | 30sec | 3min 20sec | 30 |
Liquid Culture
| Sample | medium |
|---|---|
| 9/4 RBS-lysis1+DT 3 | plusgrow(+CP) 3ml 37°C |
| 9/4 RBS-lysis2+DT 1 | plusgrow(+CP) 3ml 37°C |
| 9/4 RBS-lysis3+DT 3 | plusgrow(+CP) 3ml 37°C |
 "
"