Team:GeorgiaTech/RBS Primer
From 2013.igem.org
Most of the BioBrick parts require a Ribosomal Binding Site to express the protein. This has proven problematic as the RBS is very small and hard to insert using 3A Assembly. It has been found that the part does not even show up on a gel from gel electrophoresis, and it is therefore very hard to confirm that the RBS was inserted without sending the plasmid off for sequencing. There needs to be a more efficient way to insert a strong RBS.
We believe the creation of an oligo primer with a strong Ribosomal Binding Site, BBa_B0034, is easier and more efficient to place the RBS in front of a protein. Then, there is only a PCR reaction to be run, and the product can be digested and ligated onto a standard backbone. An easy oligo primer that could be used in PCR extension and then placed into a vector backbone would save time and frustration as to whether the part had been inserted.
The RBS primer has been optimized for use with BioBricks compatible with BioBrick standard 10. It also can only be used with parts that start with ATG, the parts with the most common need for an RBS.
Contents |
Design
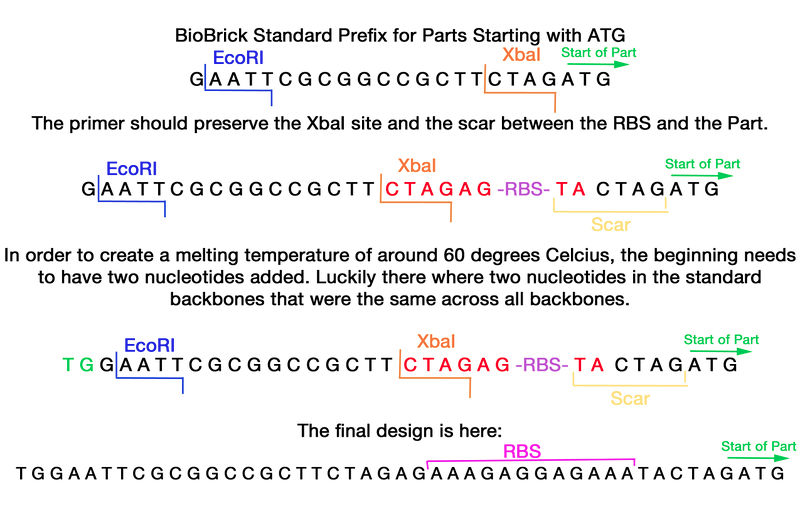
The format of this primer needs the BioBrick formatted prefix with an additional strong Ribosomal binding site within it. The prefix of the standard BioBrick was analyzed and the primer was found to be able to be inserted after the XbaI site and still have the scar intact between the RBS site and the part.
TGGAATTCGCGGCCGCTTCTAGAGAAAGAGGAGAAATACTAGATG
Procedure
The primers to use are the designed primer, and the reverse primer for BioBrick Sequencing, BBa_G00101 (VR)
The PCR Protocol is:
- 90°C for 2 minutes
- 98°C for 30 seconds
- 48°C for 30 seconds
- 68° C for 5 minutes
- Repeat steps 2-4 for 32 cycles
- End with 68°C for 10 minutes
The recipe is:
- 5x reaction buffer - 10 µL
- oligo1 (5, 10, 20 µM solution) - 7.5 µL
- oligo2 (5, 10, or 20 µM solution) - 7.5 µL
- dNTP mix (10mM each) - 1 µL
- DMSO - 1 µL
- H2O - 22.5 µL
- Phusion DNA polymerase - 0.5 µL
Put reaction in 2 tubes - negative control and reaction (50 µL each); add 0.5 µL of DNA (0.1 ng) to the reaction
Daily Progress
The primer has been ordered and should be arriving soon.
July, 12 2013
The primer has arrived and several tests will be performed to determine if the primer is usable.
I have found an RFP BioBrick part that begins with ATG, which is needed for this primer to work, BBa_E1010. I will transform and purify another part to use as a positive control, BBa_E0430.
Through these I can set up the PCR reaction to test the addition of the RBS to the BioBrick part. The primer will be diluted to 5uM, 10 uM, and 20 uM. The BioBrick for the addition of the primer will be diluted to 0.5 ng/uL. The other primer that I will use is the complementary primer for the plasmid amplification, SB-prep-2Ea.
Edit: I have now switched to using the reverse primer for BioBrick Sequencing, BBa_G00101 (VR)
The PCR Protocol will be as follows:
- 90°C for 2 minutes
- 98°C for 30 seconds
- 48°C for 30 seconds
- 68° C for 5 minutes
- Repeat steps 2-4 for 32 cycles
- End with 68°C for 10 minutes
Cells will be transformed and then purified and sent off for sequencing.
The recipe is:
- 5x reaction buffer - 10 µL
- oligo1 (5, 10, 20 µM solution) - 7.5 µL
- oligo2 (5, 10, or 20 µM solution) - 7.5 µL
- dNTP mix (10mM each) - 1 µL
- DMSO - 1 µL
- H2O - 22.5 µL
- Phusion DNA polymerase - 0.5 µL
Divide reaction in 2 tubes - negative control and reaction (~25 µL each); add 0.5 µL of DNA (0.1 ng) to the reaction
July 23, 2013
Using the 7.5 uL of the 5 uM oligo and the 363 BioBrick ASP, amplification was attempted again. This time with RFP. Results showed that RFP was amplified around the 800 bp mark, where it should be.
The amplicon was DpnI digested and PCR cleaned up with the Qiacube.
July 24, 2013
Attempted 2 ligations with RFP amplicon. 1 onto a Kanamycin backbone and the other with a pLac in front of it using 3A. These were plated on Kanamycin plates.
July 25, 2013
Growth was show on the plates, however, I decided not to do a colony PCR as the results from colony PCRs have been misleading lately.
Media solutions of 3 clones from each plate were made to be grown up overnight.
July 26, 2013
The media solutions were purified and ran a restriction digest. The results were promising although not definite. Will send off for sequencing.
July 30, 2013
Sequencing was returned. None of the clones had the RBS or any part for that matter. Strange though, some lost their XbaI site and did not have a PstI site.
July 31, 2013
Attempted the amplification again, this time with multiple different vectors that began with ATG. If the primer works, there should be bands around the respective differently length vectors. The vectors chosen were lacZa (234 bp), RFP(681 bp), and Antigen 43 (3000bp).
August 1, 2013
The next step was to gel purify each of the inserts. However, when the 0.6% gel was run, the wells made were misshapen, causing irregular bands to form. The purification was attempted regardless, but most likely this will need to be redone on new gels.
September 12, 2013
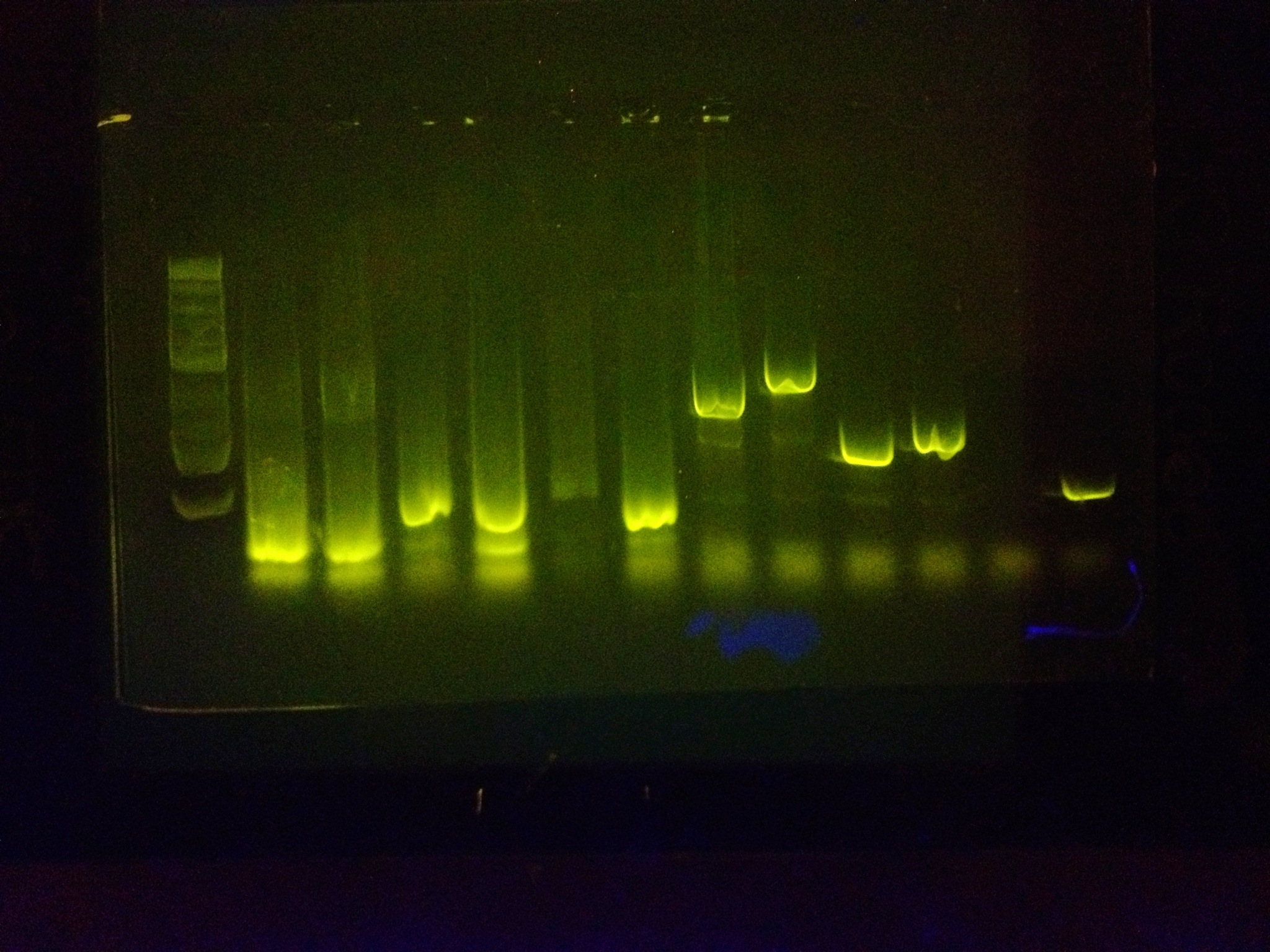
After extensive testing, many PCR reactions were run with the primer vast amount of BioBricks in unison to prove that it can amplify many BioBricks at once and be an effective tool.
Examples:
All bands beside the ladder are biobricks that have been amplified with the RBS primer and the reverse sequencing primer.
 "
"