Team:UChicago/Plan
From 2013.igem.org
Summer Plan
Construct pUB110 BioBrick
-Digest pUB110 w/ NdeI and AflII--> gel purify-->nanodrop for concentration
-Digest pUB110 linker w/ NdeI and AflII
Linker sequence:
attcgtcttaaggaattcgcggccgcttctagagtactagtagcggccgctgcagcatatgtcatac
-Set up overnight ligation of digested, gel purified pUB110 and digested pUB110 linker = generates pUB110 backbone
-Transform into B. subtilis to amplify
-Set up O. N. cultures
-Do B. subtilis miniprep
-Digest our pUB110 BioBrick
To test pUB110 backbone, which is kanamycin resistant, send for sequencing
-Put an upstream BioBrick (in vector with chloramphenicol resistance) + RFP BioBrick in an amp resistant backbone by doing 3 step assembly
-RFP expression from our modified pUB110 backbone would test if the backbone has all components needed for expression in B. subtilis
-Do transformation in B. subtilis
-Choose transformants--> if transformants express RFP, we could conclude our modified pUB110 backbone works--> submit pUB110 backbone to iGEM HQ
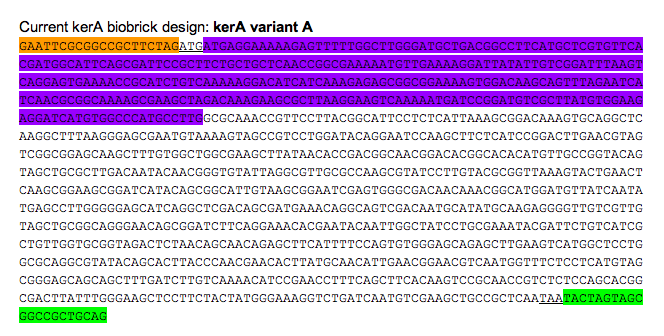
Construct kerA BioBrick
-Do Gibson assembly to put together the two kerA gBlocks from IDT
-Put our kerA biobrick into a backbone w/ amp resistance (so we can use the 3 step assembly) and transform into DH5-a
-Since promoter/upstream biobrick will be in chloramphenicol resistant vector and our puB110 vector will use kanamycin resistance, we will put the kerA biobrick in an amp resistant backbone
Orange: prefix Green: suffix Purple: kerA signal peptide
 "
"