Team:GeorgiaTech/Split GFP
From 2013.igem.org

Main Page

Split GFP
The split GFP project was one left behind by the last year’s Georgia Tech iGEM team. The idea is to take the green fluorescent protein genes and split it into two subunits (the N and C terminus), that when close enough would interact to produce GFP. This would be useful in any situation where two subunits must be close enough to interact and to determine how much interaction was needed. For this part, we began by sequencing the parts to see what might have gone wrong. One part was not BioBrick formatted and contained in EcoRI restriction site in the middle of the sequencing. Also, both needed an ATG start codon for the sequence to work. Then, the parts had to be assembled with a promoter, operator, and RBS. We used standard assembly to assemble the RBS part BBa_B0034 onto the each GFP part. We found that 3A assembly did not work for small parts such as the strong ribosomal binding site, so we had to use the standard assembly instead. Simultaneously, we assembled the strong T7 promoter (BBa_I719005) with a LacI, IPTG induced operator (BBa_R0010) using 3A assembly. After, we worked with 3A assembly to combine the T7+LacI+RBS+N-Terminus Split GFP+C-Terminus Split GFP.
Purpose
To assemble split GFP into BioBrick Vectors
Background
The 2012 GT iGEM team created a BioBrick that would allow for two parts of the green fluorescent protein to interact and allow the E. coli to fluoresce. This part needs to be assembled with T7 promoter region and a lacI operator as well as a RBS.
Starting Materials
- Split GFP vectors from the previous year
- iGEM Kits
- XL10 Gold
- XL Blue
Design
- Assemble a RBS in front of each split GFP Part
- Assemble T7 and LacI
- Assemble the T7, LacI, RBS, Split GFP BioBrick
- Transform into BL21 De3 E. coli cells that produce T7 polymerase
- Flow cytometry to determine amount of split GFP on E. coli cell
Results
June 23, 2013
1C5 - RBS+1D2. Sequencing results show that there is no part. Simply a backbone folded onto itself.

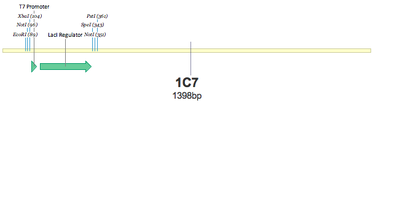
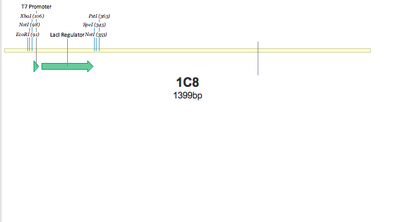
SUCCESS! Both 1C7 and 1C8 show that the T7 Promoter+lacI regulator was successfully put together using 3A Assembly.
Conclusion: Oligo Duplex does not work. Need to resuspend dry DNA biobrick BBa_B0034 in Kit Plate 5. Transform into a cell and purify the DNA for 3A Assembly.
June 26, 2013
3A Assembled BBa_B0034 RBS to split GFP parts.
June 27, 2013
Low growth on the plates. Most likely the Assembly did not work. We will run a colony PCR.
Colony PCR shows nothing but primer dimers. Insert was not in.
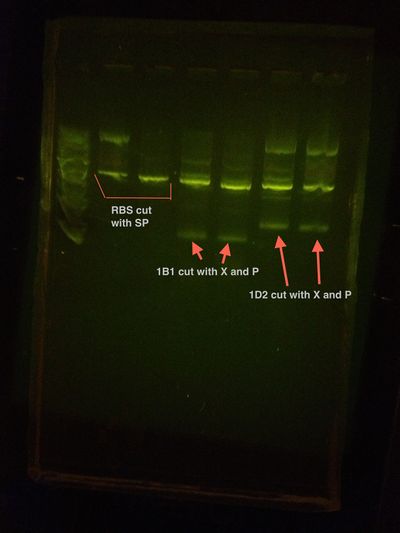
1B1 and RBS assembly Colony PCR Gel
1D2 and RBS assembly Colony PCR Gel
Conclusion: the basement machine does not work. We will try again tomorrow with upstairs thermocycler.
July 1, 2013
Colony PCR was performed on the RBS+1B1 colonies left, 10 total, that were grown over the weekend.
The results are summarized in the gels found below:
RBS+1B1 1-5 and the positive control
RBS+1B1 6-10 and the negative control
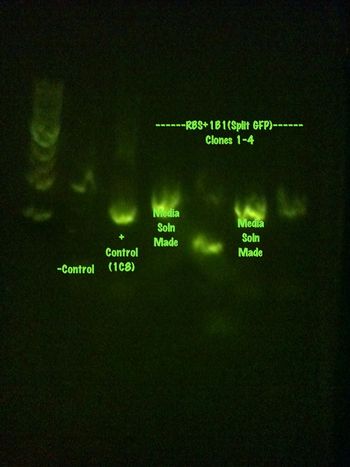
Media solutions have been created and will be grown and sent for sequencing tomorrow.
July 2, 2013
A purification of the media solutions was made using the qiacube and the gel results are shown:
It is weird that the parts cut with one restriction enzyme have three bands. I have no idea why.
July 8, 2013
The sequencing results were analyzed from macrogen. They show that there was no RBS addition.
Work on standard assembly was done, using Spencer Standard Assembly Protocol
One deviation that occurred was the enzymes ran for an hour, not an hour and a half
The gel from the enzyme digest before getting cut out and purified looked like this:
It should be noted that although the ladder was not the most clear, the backbones all lined up as they should (They are all around the same length) and the parts are around their respective lengths.
July 9, 2013
Ligation was performed, using a modified version of the 3A ligation procedure. The vectors and the parts were added at a 1:4 ratio, as that is the ratio of their relative lengths. the vector was added with 50 ng, and the part was added at 100 ng. The amount of DNA ligase and reaction buffer changed as the reaction became a 30 uL reaction, as opposed to a 20 uL reaction. 3 uL of buffer and 1.5 uL of Ligase was added instead of the usual amounts, and water filled the tube to be 30 uL.
XL Blue were transformed, since XL10 gold has been shown to be contaminated with ampicillin. The plates will be checked tomorrow for growth.
July 10, 2013
There was no growth on either of the plates. A gel confirmed that somewhere between gel purification and ligation the DNA was degraded. Another digest was run, but has yet to be purified since the QIAcube has become inaccessible.
July 15, 2013
Restriction digest and gel extraction was retried with promising results. A gel confirmed that one of the RBS vectors and a split GFP part, 1D2, were successfully purified. A ligation was performed and cells were transformed with the product.
The restriction digest was run with as much DNA as i could get from a single clone, using up almost all of a vector's stock and adjusting the amounts accordingly.
The gel that was extracted from looked like this:
July 16, 2013
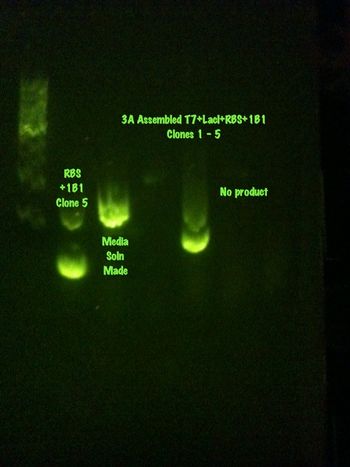
With a large amount of growth on the plates, it appears as if the assembly worked. A colony PCR was performed and it showed promising results for four of the clones. These clones have been made into media solutions and will be sent off for sequencing.
July 18, 2013
Performed Standard[3A Assembly with T7,LacI + RBS and Split GFP parts on a Cm Backbone. Casey
A standard assembly was performed with the other split GFP part using Spencer Standard Assembly Protocol and is awaiting a gel to confirm the purification of the gel extraction. This was performed with the same modification of the steps, including using all of the DNA from a single clone in the restriction digest to attain the largest amount of concentration on the gel to extract.
July 19, 2013
The sequencing result came back from Macrogen today, and the assembly was a success! It was proven to work with split GFP part 1D2, BBa_K916003.
Work has began on combining the other part, 1B1, BBa_K916004.
Also began the 3A Assembly of T7+LacI with the new RBS+1D2. However there was a mistake using ampicillin backbone when the chloromphenicol should have been used instead
July 22, 2013
Colony PCR of the products were ran.
The colony PCR also came out strange. Possibly the gels made this morning were not the best batch. 1B1 clones 1 and 3 were made into media solutions. It appears there was a slight contamination around the 1kb mark so any faint marks at this level were disregarded as contaminants. None of the 3A Assembled products appear to be the right size (800 bp). This is most likely to do with my mistake in the 3A assembly. I should've used a chloromphenicol backbone for the proper 3A Assembly. Clone 1 appears to be the most promising and will be made into a media solution.
August 1st, 2013
Sequencing came back for the assembled T7, LacI, RBS, and 1D2, which shows nothing. Another 3A Assembly will be attempted today for both 1D2 and 1B1.
August 7th, 2013
After suffering some setbacks, including no gels to run DNA, it was determined that the chloramphenicol backbone may be incorrect. All tried assemblies onto this backbone have been unsuccessful.
September 13, 2013
3A Assembly of full operating GFP part. Colonies grew, however the colony PCR did not show the most promising bands. Will send for sequencing.
September 18, 2013
Sequencing results do not look promising. Still a lot of problems with our current chloramphenicol backbone. Must do testing to determine why the parts are not assembling well in the 3A Assembly. Maybe we should revert back to standard assembly.
 "
"