Team:Exeter/Results
From 2013.igem.org
Green Light Module
We have had marked success with our Green Light Module. Not only is the magenta pigment bright and evenly synthesised, but we have also managed to show magenta synthesis is inhibited by exposure to white light. We are currently working on showing that green light specifically switches off magenta production.
| our vibrant magenta pigment | clearly show magenta pigment | grown in dark vs. light |
Cyan pigment
We managed to get our cyan pigment working in the labs after some difficulties with our digestions/ligations. The pigment takes a while to develop, over 24 hours compared to magenta's overnight development, but the colour is reasonably strong and is produced evenly by the cells. However, more time has been invested in the cyan pigment which is responsive to red light (see "Planned Post-Freeze Work").
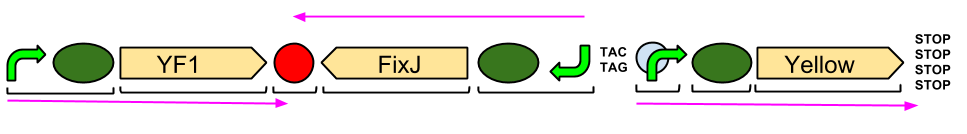
Red Light Module output Brick with BglII and BamHI restriction sites
| assembly parts showing superfolder GFP |
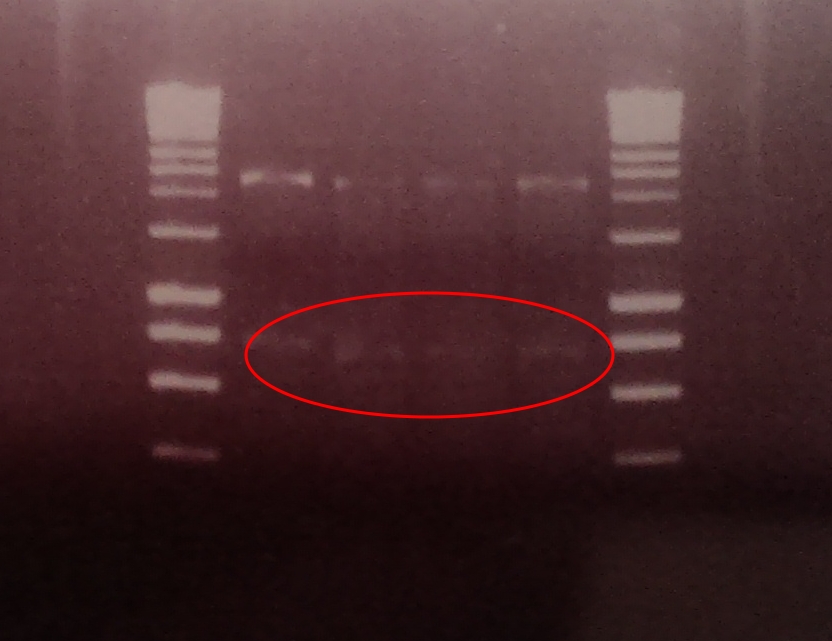
| output bricks showing band at around 950bp |
Our output brick for the Red Light Module includes the ompC promoter which is specific to the protein [http://en.wikipedia.org/wiki/Porin_response:_osmoregulation_in_E.coli OmpR]. This is followed by an RBS and a restriction site for BglII and the coding region for a [http://parts.igem.org/Part:BBa_E0040 GFP]. A second restriction site for BamHI follows, then a series of STOP codons to act as a terminator. The whole sequence is flanked by the iGEM prefix and suffix allowing the BioBrick to be used in 3A Assembly.
This BioBrick was built for us by IDT in their gBlock format. We designed the biobrick to contain a central GFP that is split between the two gBlocks, thus acting as a marker for a successful gibson assembly.
As the image shows, the two halves of GFP have been ligated together to form a complete, working protein in the successful Gibson assembly of our output bricks. GFP is constitutively expressed within DH5α cells as endogenous ENVZ phosporylates endogenous OmpR as a component of osmolarity sensing. Endogenous OmpR then binds to ompC. Thus our output brick is working as expected. In order to test whether the output brick can be implemented into the Red light pathway and ENVZ- strain should be used to avoid false positives.
A restriction digest was performed on 4 independent Gibson assembly products with BamHI and BglII to ascertain whether the restriction sites were implemented correctly into our brick. A gel electrophoresis of the digested parts showed two clear bands: plasmid DNA with ompC promoter and suffix at ~2300, and superfolder GFP coding sequence DNA at ~750. Both bands are in their expected position.
In order to insert a new part, we suggest following a standard primer based mutagenesis protocol to change the restriction sites to BamHI and BglII. Additionally when picking transformed colonies, screen for green fluorescence. These colonies' plasmids have ligated incorrectly. Colonies' plasmids that have no florescence should be miniprepped, digested and ran on a gel to check for the correct insert.
Planned "Post-Freeze" Work
Blue Light Module
Our Blue Light Module, which is being built by DNA2.0, will unfortunately not arrive in the labs until after the Wiki-freeze. However, we will be bringing plenty of pictures of it working (hopefully!) to the Jamboree. Considering it has been constructed in a very similar manner to the Green Light Module, which is functioning in the lab, we are reasonably hopefully that we can have the Blue Light Module functioning in the lab soon after its arrival.
Red Light Module
We have successfully attached the OmpR specific promoter ompC to the cyan pigment coding region, and are now working on transforming the plasmid into EnvZ deficient cells (kindly provided to us by the Dundee iGEM 2013 Team).
This is due to OmpR and ompC being naturally present in most E. coli competent cells as part of this osmoregulation. EnvZ is the transmembrane protein which phosphorylates OmpR (to become OmpR-P) when osmolarity in the cell changes; its phosphorylation triggers the production of outer membrane porins. In our project, we only want OmpR to be phosphorylated when the cells are exposed to green or blue light, and for phosphorylation to halt when exposed to red light. Obviously, if OmpR-P is being formed outside of these rules, we will be getting pigment production (or lack thereof) when we have not allowed it, which will ruin the images we are trying to develop. Hence, we will express the Red Light Module in EnvZ deficient cells. The plasmid for the Red Light Module contains coding for EnvZ, but also for the second half of the transmembrane protein which allows our system to be light responsive; Cph1. EnvZ and Cph1 work together to form Cph8.
 "
"