Team:Bielefeld-Germany/Project/Nanowires
From 2013.igem.org
Nanowires
Overview
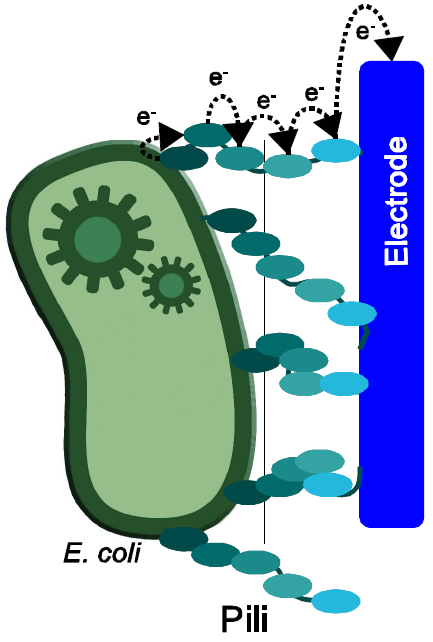
Multiple bacteria form special, highly electrically conductive pili, which are required for survival in anaerobe environment. Electrons, which are generated through the oxidation of different substrates, can be transported by these pili and transferred to alternative solid electron-acceptors, such as sulfur or iron compounds. These properties make them another interesting option, regarding the optimization of E. coli for the use in MFCs especially because the so called nanowires could increase the number of bacteria contacting the surface of the anode. Unfortunately multiple genes, ordered in large gene clusters are required to form nanowires in organisms such as Geobacter sulfurreducens, so that the cloning of a functional expression system seems to be a big challenge. For that reason the genetic modification of the existing non-electrically conductive type 4 pili from Escherichia coli is another very interesting option to utilize E. coli as a MFC-organism.
Theory
Next to the well-known group of pili-structures, which are responsible for conjugal gene transfer and called F-Pili, several other types of these hair like structures can be found, which accomplish a bride range of different purposes. The pilus-classification is based on specific host-receptor interactions, verifiability with different antibodies and the amino acid sequence of the pilin protein, which is the conserved oligomeric element, found in every pili-structure (Strom and Lory., 1997). One of these families, containing the so called Type IV pilis, is very interesting for building a microbial fuel cell, because some of these protein structures are able to conduct electrons. In some anaerobic habitats bacteria depend on the electron transfer to electron acceptors like Fe(III)oxides to evolve the electrons, generated during their metabolic activities. The insoluble condition of such compounds selected for a complex network of conductive structures, which electrically connect the bacteria with their environment (Shi et al., 2007). Based on these properties the use of biocompatible electrodes could enable the bacteria to execute anaerobic respiration with a simultaneous usage of the released electrons for the production of electricity.
In microbial fuel cells, filled with waste water and inoculated with sewage sludge under anaerobic conditions a formation of a thick biofilms is observed for the anode, which contains a mixed culture of typical organisms. Next to species like Shewanella putrefaciens and Rhodoferax ferrireducens mainly representatives of the genus Geobacter are found, which are capable of using an electrode as their final electron acceptor (Kim et al., 2005). In particular Geobacter sulfurreducens is well known for its possibility to reduce Fe(III)Oxides and other insoluble compounds by formation of cytochromes in the outer membrane and production of electrically conductive pili, which are also called nanowires. Due to the fact that many studies have been conducted to evaluate the special properties for this group of bacteria used Geobacter sulfurreducens it can be defined as a model organism. Not least because of its genetic tractability and the fact, that it’s one of the first fully sequenced organisms of this group it stands to reason to use the pili formation gene complex from this species to modify the target organism Escherichia coli with regard to the production of electrically conductive pili (Coppi et al., 2001).
In contrast to the use of mediators, which cause an indirect electron transfer the use of nanowires could enable Escherichia coli to use direct anaerobic respiration without limiting effects like diffusion or the electron transfer between redox-mediator and anode. With respect to the use of cytochromes, which could render anaerobic respiration by direct cell-anode contact, too, nanowires could allow a much more bacteria to contact with an electrode because of the surface enlarging effect of the protein extends.
The proposed usage of genetically engineered Escherichia coli Type IV pili, by changing the amino acid sequence with regard to an increased contend of aromatic amino acids could not be realized. Although this modification was very promising, since further studies clarified the importance of these amino acid type for long range extracellular electron transfer in Geobacter sulfurreducens, a implementation of the project was not possible because of the pathogenicity association of all existing type IV pili from Escherichia coli (Vargas et al., 2013). This fact renders this approach very uninteresting for a usage as the part of a microbial fuel cell system, which should be free from any safety problems, with the result that it was not further considered.
Genetic Approach
In general, all pili-like structures consist mainly of oligomeric pilin-proteins. For Geobacter sulfurreducens the corresponding gene is called PilA and includes 273 nucleotides. Previous studies spelled out, that next to this central gene at least one big cluster of other genes is essential for functional expression of the Geobacter nanowires, by complementing a pilA knockout by means of a plasmid located pilA gene. Without the entire open reading frame GSU 1496-1505 it was not possible to complement the knockout in view of a functional nanowire expression. Furthermore the genes pilB, pilT, pilC, pilS and pilR carry out important tasks in relation to a functional pilA expression, like the regulation of pilA transcription (Richter et al., 2012). The relevant open reading frame is illustrated in figure 2.
Because of its capital size of 16362 bp, the gene cluster GSU 1491-1505 was devided into two big parts of about 7,2 and 9 kb to permit the use of the gibson assembly. This is essential to turn the cluster into the Biobrick form, because the containing forbidden restriction sites for EcrRI and PstI hinder the use of the standard Biobrick assembly. In addition further studies showed, that the G. sulfurreducens genome sequence owns two functional translation start codons for pilA, resulting in two isoforms of the PilA protein which are both essential for the formation of functional nanowires (Richter et al., 2012). In this contend two different version of the open reading frame GSU 1496-1505, differing in the existence of the natural G. sulfurreducens promoter were used to analyze its functionality in Escherichia coli.
Because the alignment of all fifteen genes illustrated no equivalent genes in the Escherichia coli genome, the insertion of the whole fragment to the high copy plasmid pSB1C3 is necessary and should enable E. coli to produce functional nanowires.
Since there were no information available, whether the pilin secretion system of the type IV pili producing E. coli strains still exists in the appropriate stain E. coli KRX another gene Geobacter sulfurreducens open reading frame, containing the respective secretion proteins, could be relevant for a functional expression (Reguera et al. 2005). The gene structure of this cluster including 9042 bp is shown in figure 3.
Results
After the successful amplification of the Geobacter sulfurreducens clusters GSU 1491-1495, GSU 1496-1505 and GSU Promoter 1496-1505 the transformation of these long DNA sequences into the Biobrick form, using the Gibson assembly, lead to substantial problems. Not at least because of the already explained inaccessibility to the standard Biobrick assembly and the not very promising prospects for a functional expression of nanowires in Escherichia coli, resulting from the big number of essential proteins and the complex promoter-structure, the nanowire project was stopped under further notice to focus on the other projects.
References
Coppi, M. V., Leang, C., Sandler, S. J., & Lovley, D. R. (2001). Development of a Genetic System for Geobacter sulfurreducens. [http://aem.asm.org/content/67/7/3180.short Applied and environmental microbiology, 67](7), 3180-3187.
Kim, J. R., Min, B., & Logan, B. E. (2005). Evaluation of procedures to acclimate a microbial fuel cell for electricity production. [http://link.springer.com/article/10.1007/s00253-004-1845-6 Applied microbiology and biotechnology, 68](1), 23-30.valuation of procedures to acclimate a microbial fuel cell for electricity production.
Reguera, G., McCarthy, K. D., Mehta, T., Nicoll, J. S., Tuominen, M. T., & Lovley, D. R. (2005). Extracellular electron transfer via microbial nanowires. [http://www.nature.com/nature/journal/v435/n7045/abs/nature03661.html Nature, 435](7045), 1098-1101.
Richter, L. V., Sandler, S. J., & Weis, R. M. (2012). Two isoforms of Geobacter sulfurreducens PilA have distinct roles in pilus biogenesis, cytochrome localization, extracellular electron transfer, and biofilm formation. [http://jb.asm.org/content/194/10/2551.short Journal of bacteriology, 194](10), 2551-2563.
Shi, L., Squier, T. C., Zachara, J. M., & Fredrickson, J. K. (2007). Respiration of metal (hydr) oxides by Shewanella and Geobacter: a key role for multihaem c‐type cytochromes. [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2958.2007.05783.x/full Molecular microbiology, 65](1), 12-20.
Strom, M. S., & Lory, S. (1993). Structure-function and biogenesis of the type IV pili. [http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.47.100193.003025 Annual Reviews in Microbiology, 47](1), 565-596.
Vargas, M., Malvankar, N. S., Tremblay, P. L., Leang, C., Smith, J. A., Patel, P., ... & Lovley, D. R. (2013). Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. [http://mbio.asm.org/content/4/2/e00105-13.short mBio, 4](2).
 "
"