 The Riding
The Riding
 The Riding
The Riding
The first objective of our project was to genetically engineer bacteria so they can be able to “ride” the worms. Bacteria are not able to move fast in
through solid or semi-solid substrates but they are very interesting from a biotechnological point of view, that is the reason why we though in this
innovative mean of transport.
To achieve our goal, we constructed a BioBrick (see part: BBa_K1112001)
consisting in the coding sequence of the hmsHFRS operon, an adhesion operon natural from Xenorhadbus nematophila which allows the formation of a
biofilm on the nematode S. carpocapsae, under the control of a nitrogen sensitive promoter (pGlnA, characterized by the 2012 Valencia Biocampus iGEM
team).
Our original idea was to transform Pseudomonas putida with the BioBrick, but the efficiency of the transformation was too low, so we decided to try
with E. coli. Pseudomonas transformation with the operon could not be directly achieved with the vector in which the BioBrick was synthesized
(pUC57 vector) so that we tried to insert the BioBrick in a vector with origin of replication in both Pseudomonas and E.coli, the pIZ1016
destiny vector (6,5Kb).
The mechanism of the construction works as follows: with low nitrogen in the media (0,6 g/L), the promoter is activated and hms genes are expressed,
then the biofilm formation triggers; With high nitrogen concentrations (NH4(SO2)), as it is the case of nutrient-rich hotspots, the
promoter is repressed and Pseudomonas should get out of the nematode. This process is what we call the riding.

Figure 1. Schematic representation of the BioBrick used for the riding. The effect of nitrogen concentration is shown in the drawings.
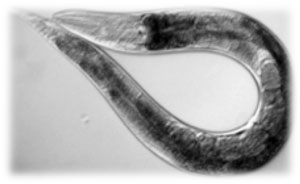
As you can see in the following electron micrography E.coli is able to colonize C.elegans’ cuticle under specific conditions:

 The Calling
The Calling
 The Calling
The Calling
Looking for an attractant
When we were considering setting up a transport of bacteria was thought to be necessary to have a 'destination', a place to go. That destiny would be a
'hot spot' on a heterogeneous substrate where
Caenorhabditis elegans should lead right to that point the bacteria.
Leveraging the powerful smell of the nematode, it was decided to try a number of attractants from various lists from web
www.wormbook.org that could work as 'hot spot' of our experiment. Thus, using the
C. elegans chemotaxis, we could direct transport.
The attractants experiment
The test would be carried creating our own plates on which half would be NGM unmodified and the other half part would be including soluble compounds before
solidifying or after solidification in the case of volatiles. The list of modifications can be found in
Fig. 1.

To place C. elegans on the plate, different cuts were made on a fresh plate of NGM, the resulting small pieces were placed in the exact
center of the 50% -50% plates to determine which side of the nematode preferred, one per Petri plate.
The results after counting 2 replicates per attractant can be seen in Fig. 2.

From Table 2 it could rule out many of the attractants that were thought viable.
Volatile attractants were not a good choice to evaporate quickly (which is also limited to the field experiments).
Another impediment arose. Once had already performed the experiments, it was decided that the promoter that will control the production of RNA interference
in Escherichia coli would be controlled by nitrogen, amino acids had to be discard as attractants; it would modify controlled expression of E.coli.
That left the MgSO4 and hypoosmotic media as potential attractants.
Attractant final choice
Because the hyposmotic medium could interfere with the proper growth of bacteria (food of our nematode), the final decision was to choose the MgSO
4 as attractant.
The question that arose at that time was: How we can multiply the amount of MgSO4 to increase efficiency?
MgSO4 efficiency
Once we had selected the most feasible attractant to the ‘transport’ for our experiments, we needed to know what could be the largest concentration of MgSO
4 in order to optimize the attraction of the nematode.
To choose the concentration, were tested in a battery of increased concentrations regarding the initial medium (1 ml / L). Bearing in mind the results of
the factor x2, we decided to test other factor concentrations: x3, x4, x5, x8 and x10; covering a range that does not exceed the concentration at which
might affect the life of
C. elegans or bacteria.
Moreover, approaching experiments with
E. coli and
Pseudomonas putida, these trials were testing the end media: half plate with NGM
non-altered and half as PHA production medium for
Pseudomonas and interference of
E. coli.
Not knowing what fatty acid could activate transcription better, we tested two possibilities: oleic acid (named in the tables as PHAol) and octanoic acid
(PHAoc). The concentration for each fatty acid was tested in
E. coli choosing as better:
- 1.28 µl/ml of Octanoic acid.
- 2.58 µl/ml of Oleic acid.
You can see our results in figures 3, 4, 5 and 6. The two first counts were made after 3 hours and the next ones after 6 hours. We suppose at that
time the worms can select their “favorite” half part gone across and their movement would be always in the same area. We prepare as control plates with the
same composition but without MgSO4 in PHA media.

Final concentration choice
Once the test already performed and that the results seen by the factor of PHA and MgSO
4 selected fatty acid give very scattered (there is
probably repellent effect at high concentrations in the medium with oleic but reversed in octanoic) we make a selection of MgSO
4 concentration
for each medium.
- If it is used oleic acid à Better results with 4ml/L MgSO4.
-
If octanoic acid used à Best results to 10 ml/L of MgSO4.
Octanoic was discarded as a transcriptional activator of the iRNA of clumping after other result of
E. coli, so finally we selected PHA medium with
oleic acid.
The biggest problem in trying to have an effective attractant was
how effective could be in presence of E. coli. It was therefore necessary
to test the tradeoff between the value of 4ml / L MgSO
4 and pair it with different concentrations of bacteria, high enough to feed the nematode
but low enough to permit the attractive effect of MgSO
4.
To find this point of commitment we prepare experiments in which the concentration of 4ml/L of MgSO
4 is faced against different ODs from serial
dilutions of a preculture of E. coli DH5a.
Table 7
shows the results. The count took place at 3h after the passing of fresh nematodes.
Best E. coli OD choice
An OD of 1 (minimum concentration of cells / volume) gives an attractive effect even better than expected (subsequent experiments try to see if there is
synergy between
E. coli and attractive factors MgSO4).
To improve the approximation was decided to repeat the experiment with MgSO
4 concentration and the chosen bacteria OD showing that the system
works well. The results can be seen in
figure 8.

 The Clumping
The Clumping
 The Clumping
The Clumping
The base of this part is our engineered E.coli with a biobrick that is capable of changing the worm’s behavior.
Our C. elegans strain (N2 strain) has two feeding behaviors: Social and solitary feeding. Social feeding is known as clumping
as you can see in this video:
Clumping is known to be induced under some conditions like temperature or starving, something that we had had considered during the entire project.
However, it is also known how clumping is controlled from a genetic perspective and the main genes have already been described. We thought we could use
this genetic approximation to control clumping under specific and controlled conditions. (view “Natural variation in a neuropeptide Y receptor
homolog modifies social behavior and food response in C. elegans.” Bono M, Bargmann CI).
How do we induce clumping?
Our initial thought was to interfere the principal gene involved in this route:
NPR-1. It is known that mutations in
NPR1 convert a solitary
strain into a social strain. Another option we finally chose was
FLP-21, a gene that positively regulates
NPR-1. We selected this one instead
because it is not involved in so many vital processes.
FLP-21 encodes a single FMRF amide-related neuropeptide that serves as a ligand for
Npr-1, a G protein-coupled receptor that regulates social versus solitary feeding behavior in several
Caenorhabditis species (see
fig. 1).

So, this would be the final process: engineered E.coli that expresses FLP-21, whom RNA will interfere with the one codified by
the worm because of the ingestion of bacteria. The formation of a complex of dsRNA will induce the elimination of Flp-21 transcripts by the RNA
silencing pathway, which consequence is the induction of clumping (see figure 2). We also had to take into account that the induction of clumping by
this mechanism is almost immediate, in comparison with natural clumping which takes longer depending on the external factors.

Why do we induce clumping for?
The main objective of the synthetic symbiosis that we designed was to detect hotspots of interest in irregular substrates and the induction of clumping was a good tool for it. With this natural mechanism, which we were going to control,
worms kept in the desired places, then
bacteria transported by C. elegans (
Pseudomonas putida) were concentrated where their action was needed, in order to generate value-added products such as bioplastic PHA and the increase of worms in a specific location
improved the image-based detection mechanism (
go to Devices section)
Controlling the mechanism
The social feeding behavior needed to be controlled; it might be induced by conditions of interest as a tool for detecting hotspots in irregular substrates. We decided that
fatty acids were going to be the inductors of the promoter (
fadBp) that regulated the expression in the FLP-21
antisense sequence, in order to induce RNA interference (
Fig.3). In the fatty acid rich media where this mechanism was activated, bacteria got off
the worm, so this construction allows the induction of clumping in the spots where a biotechnological process will be carried out.

Choosing the right fatty acid
The
E. coli’s biobrick promoter is induced by fatty acids, but those also induce the promoter of
Pseudomonas putida in order to
produce PHA. Some investigations showed that the induction
of fadBp promoter by
oleic acid is the most efficient (
view “Regulation of
fatty acid degradation in Escherichia coli: analysis by operon fusion” Clark D. et al) is
higher when the fatty acid used is
oleic acid but this is a long chained compound and that could affect the production of bioplastic (PHA). We
decided to test the growth of
E. coli and the production of bioplastic by
P. putida using PHA media with oleic acid and
octanoic acid in order to get the best results in both activities.
Both species of microorganisms where grown in PHA production media with different concentrations of oleic and octanoic acids, maintaining a global
concentration of fatty acids of 8mM, because it was the one where we found the highest growth of both E. coli and P. putida. Table 2
(below) summarizes the assays done.

The results of this assay showed that E. coli growth was better on PHA production media + 8mM oleic acid and the P. putida production of
bioplastic was acceptable in this conditions. However, the highest production of bioplastic was on PHA production media+ 8mM octanoic acid one. So then, we
made a Colonization assay and finally we determined that we were going to continue working using the PHA production media + 8mM oleic acid. Also,
the ODs for plating both organisms were stablished as a consequence of this experiment. The “Choosing the right fatty acid” and the “Colonization” assays are explained at Building the bioplastic section.
Observing clumping induced by fatty acid rich media
We could see how clumping was done induced by the presence of
8mM of oleic acid (2.58 µl/ml ) which regulated the
FadBp promoter of
E. coli and as a consequence the expression the complementary sequence to the worms’
FLP-21 transcript, producing interference
by RNA. In the plates there was a high concentration of
E. coli from the serial centrifugations preculture 4ml
XL1-Blue, 2 min at maximum rpm
(2 times). The following images show the change of behavior of
C. elegans, which at first were eating alone and then they were doing it in groups as
it was planned
(Fig. 4-6).

 The Building
The Building
 The Building
The Building
We decided to include in the mechanism design a regulated production of a value-added product in the hotspots of interest were C. elegans would be
attracted to. We got proffit of the natural capacity of Pseudomonas putida of producing bioplastic PHA (polyhydroxyalkanoates) by bacterial fermentation of
sugar or lipids (fatty acids in this case), because we found that the directed production of this versatile material was interesting due to its easy
detection (complexes of PHA with red Nile are fluorescent), its bright future in the field of biomaterials and because we were using this natural ability
of bacteria as a tool.

To regulate the expression of the PHA cluster we used the glnA promoter that our team characterized the last year iGEM’s competition. (see https://2012.igem.org/Team:Valencia_Biocampus/Results1 ) for that, we optimized
the concentration of ammonium sulphate for our Pseudomonas. We amplified the operon sequence and we sent it to the register (reference BBa_K1112002)
The role of the operon in our project
Our 2013 project is a proof of concept of the benefits that an artificial synthetic symbiosis between bacteria and nematodes can offer. The roles
Pseudomonas play are two: firstly, they have been engineered to “ride” on
C. elegans by the formation of a biofilm, but not stuffed with that, under the promoter glnA we stimulated the production of PHA, a value-added product that can be widely applied, for example:
-
For short disposable packaging items (personal hygiene products, surgical clothes).
-
In upholstery.
-
In the photographic and printing industry
-
For textile industry since PHA can be processed into fibers.
-
For biofuel production from PHA obtained from sewage sludge. This would combine two major advantages: the wastewater treatment and the generation of
energy.
Choosing the right media
On the plates we were going to have both
P. putida and
E. coli and their promoters’ response towards fatty acids was different, so we had to
make a media where
E. coli could induce
clumping and
P. putida the
production of bioplastic. In
E. coli, the
briobrick promoter for the expression of
FLP-21 iRNA engineered in order to induce the social feeding
behavior of
C. elegans (Clumping) is induced by fatty acids, but not all the fatty acids are able to induce the same level expression
(
Fig. 2);
oleic acid produces the highest activation of the promoter.

Whereas the standard medium for the production of PHA by Pseudomonas putida (composition reflected in Fig.3) employs octanoic acid (a short-chain fatty acid) as inducer. This fact made necessary to check if the modification of the fatty acid used, octanoic by
oleic, would modify the bioplastic (PHA) production.

Otherwise, the medium for PHA production is tuned for the development of Pseudomonas putida, but not for the E.coli, so we had to test if
this bacteria was able to grow in it. When we did this checking, we realized that the development of these bacteria did not occur in the PHA production
media. However, with the addition of acetate (carbon source), the removal of the iron salt (added to the media to avoid the synthesis of a siderophore by Pseudomonas sp.) and, due to serendipity, with the double
concentration of trace elementes, we got the media for the enteric bacteria growth.
Choosing the right fatty acid
Firstly, we tried to grow
E. coli in the new PHA production media with different concentrations of octanoic acid. The same assays were done in
parallel with
P. putida in order to check which were the conditions where both microorganisms could grow and
P. putida could produce enough
quantity of bioplastic.
After overnight incubation, the cultures were washed using sterile PBS and ODs600 were measured (Fig.4.). We also checked the PHA production by centrifuging the precultures, resuspending the pellet on PBS and adding Red Nile dissolved in DMSO, we could see the
emission of fluorescence due to the interaction between the Nile red and the PHA through an UV transilluminator (Fig.5). By the results of the
assays we chose to work with a 8mM concentration of fatty acids (Fig.4).

We also checked the PHA production by centrifuging the precultures, resuspending the pellet on PBS and adding Red Nile dissolved in DMSO, we could
see the emission of fluorescence due to the interaction between the Nile red and the PHA through an UV transilluminator (Fig.5). By the results of
the assays we chose to work with a 8mM concentration of fatty acids (Fig.4).

At this point we found that E. coli and P. putida could grow at 8mM of octanoic acid and that bioplastic (PHA) is also produced. However,
because E. coli grows better in oleic acid and we wanted to check if P. putida could produce PHA if its phaC operon was induced
by it, we grew both bacteria in different concentrations of octanoic and oleic acid but maintaining a constant concentration of fatty acids of 8mM.
The assays done are explained in the table below (Fig.6).

We saw that E. coli had grown better with the increasing of oleic acid, getting at its maximum on experiment 9, even so, the quantity of pellet
corresponding to 1 mL of preculture was scarce (and not visible at experiments 1 to 5). E. coli needed more time to grow in the PHA production
media.
P. Putida
had grown too, however we wanted to measure the production of bioplastic, we measured the ODs600 from the precultures of P. putida by
centrifugating 2 mL of them and then washing them on 1 mL of PBS and resuspending the pellet in 500 μl of PBS. Then, dilutions on PBS were
adjusted to the lowest OD600,corresponding to the experiment number 9 (8mM oleic acid). The growth of P. putida lower in PHA
production media with a concentration of 8mM of oleic acid (Fig. 7-8).

The results showed that the growth of E. coli on 8mM of oleic acid was better than in any other condition, then experiment 9
conditions would be the best ones for the induction of clumping. However, even P. putida had produced bioplastic at all oleic acid
concentrations, the highest production levels corresponded to experiment 1 conditions, which means in absence of oleic acid, just on 8mM of octanoic
acid. Because of that, next assays were done following the conditions of experiment 1 and 9 (Fig.9).

After the optimization of the media, kinetics of the production of PHA and biomass generation were realized in relation to the time (Fig.10).

Colonization assays
For establishing the right media, the one in which
E. coli could be able to grow and induce clumping and
P. putida produce PHA and get to a
balance
between the two activities and for setting up some standard bases for doing an assay with the three microorganisms chosen for this synthetic
symbiosis that our team proposes, we made some colonization assays. Different ODs
600 of
E. coli where plated in PHA production media in
the conditions of both
experiments 1 and 9 (
Fig.6). There were two sets of plates:
-
1) PHA production media + 8mM oleic acid
-
2) PHA production media + 8mM octanoic acid
In both of them (and their replicas) were plated the following E. coli ODs600 (Fig.11) and after let them grow for 48 hours, the
plates were divided into 4 parts in order to plate different P. putida ODs600 (Fig.11). The huge difference between the ODs chosen
for E. coli and P. putida was because we wanted to plate similar quantities of the Pseudomonas that C. elegans could afford to
carry, taking into account that the worm size is 1mm, approximately.

Then the production of bioplastic was seen by the fluorescence of the Nile red joined to the PHA (the samples were prepared as in The right media).
There were taken pictures of the plates but there was only growth of Pseudomonas in the first set of plates (PHA production media + 8mM oleic acid).
There was a notorious growth and production of bioplastic in the plates where E. coli was plated with an OD of 0,8 an 1,0 and the production of
bioplastic was higher at 0,03 and 0,05 OD600 of P. putida. (Fig.12).





 "
"
























